The AMPK-related kinase NUAK2 suppresses glutathione peroxidase 4 expression and promotes ferroptotic cell death in breast cancer cells
- PMID: 35523770
- PMCID: PMC9076840
- DOI: 10.1038/s41420-022-01044-y
The AMPK-related kinase NUAK2 suppresses glutathione peroxidase 4 expression and promotes ferroptotic cell death in breast cancer cells
Abstract
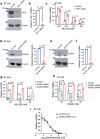
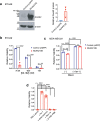
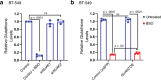
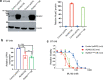
Ferroptosis is a caspase-independent form of regulated cell death strongly linked to the accumulation of reactive lipid hydroperoxides. Lipid hydroperoxides are neutralized in cells by glutathione peroxidase 4 (GPX4) and inhibitors of GPX4 are potent ferroptosis inducers with therapeutic potential in cancer. Here we report that siRNA-mediated silencing of the AMPK-related kinase NUAK2 suppresses cell death by small-molecule inducers of ferroptosis but not apoptosis. Mechanistically we find that NUAK2 suppresses the expression of GPX4 at the RNA level and enhances ferroptosis triggered by GPX4 inhibitors in a manner independent of its kinase activity. NUAK2 is amplified along with MDM4 in a subset of breast cancers, particularly the claudin-low subset, suggesting that this may predict vulnerability to GPX4 inhibitors. These findings identify a novel pathway regulating GPX4 expression as well as ferroptotic sensitivity with potential as a biomarker of breast cancer patients that might respond to GPX4 inhibition as a therapeutic strategy.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

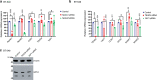
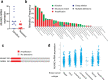
Similar articles
-
Ferroptotic cell death triggered by conjugated linolenic acids is mediated by ACSL1.Nat Commun. 2021 Apr 14;12(1):2244. doi: 10.1038/s41467-021-22471-y. Nat Commun. 2021. PMID: 33854057 Free PMC article.
-
Metadherin enhances vulnerability of cancer cells to ferroptosis.Cell Death Dis. 2019 Sep 17;10(10):682. doi: 10.1038/s41419-019-1897-2. Cell Death Dis. 2019. PMID: 31527591 Free PMC article.
-
xCT-Driven Expression of GPX4 Determines Sensitivity of Breast Cancer Cells to Ferroptosis Inducers.Antioxidants (Basel). 2021 Feb 20;10(2):317. doi: 10.3390/antiox10020317. Antioxidants (Basel). 2021. PMID: 33672555 Free PMC article.
-
GPX4 at the Crossroads of Lipid Homeostasis and Ferroptosis.Proteomics. 2019 Sep;19(18):e1800311. doi: 10.1002/pmic.201800311. Epub 2019 May 31. Proteomics. 2019. PMID: 30888116 Review.
-
Lipid Peroxidation-Dependent Cell Death Regulated by GPx4 and Ferroptosis.Curr Top Microbiol Immunol. 2017;403:143-170. doi: 10.1007/82_2016_508. Curr Top Microbiol Immunol. 2017. PMID: 28204974 Review.
Cited by
-
Identification of biomarkers associated with ferroptosis in diabetic retinopathy based on WGCNA and machine learning.Front Genet. 2024 May 28;15:1376771. doi: 10.3389/fgene.2024.1376771. eCollection 2024. Front Genet. 2024. PMID: 38863444 Free PMC article.
-
Depletion of NUAK2 blocks the stemness and angiogenesis and facilitates senescence of lung adenocarcinoma cells via enhancing ferroptosis.Cell Div. 2024 Jul 28;19(1):23. doi: 10.1186/s13008-024-00128-8. Cell Div. 2024. PMID: 39068449 Free PMC article.
-
CircUBE3B High Expression Participates in Sevoflurane-Induced Human Hippocampal Neuron Injury via Targeting miR-326 and Regulating MYD88 Expression.Neurotox Res. 2023 Feb;41(1):16-28. doi: 10.1007/s12640-022-00617-0. Epub 2022 Dec 31. Neurotox Res. 2023. PMID: 36585543
-
Kinome profiling: A veritable Rosetta Stone for protease-activated receptor 1 biased signaling.Vascul Pharmacol. 2025 Mar;158:107469. doi: 10.1016/j.vph.2025.107469. Epub 2025 Feb 2. Vascul Pharmacol. 2025. PMID: 39904417 No abstract available.
-
Identification of TFR2 as a novel ferroptosis‑related gene that serves an important role in prognosis and progression of triple‑negative breast cancer.Oncol Lett. 2023 Dec 5;27(2):43. doi: 10.3892/ol.2023.14176. eCollection 2024 Feb. Oncol Lett. 2023. PMID: 38106522 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

