Laminar shear stress inhibits inflammation by activating autophagy in human aortic endothelial cells through HMGB1 nuclear translocation
- PMID: 35523945
- PMCID: PMC9076621
- DOI: 10.1038/s42003-022-03392-y
Laminar shear stress inhibits inflammation by activating autophagy in human aortic endothelial cells through HMGB1 nuclear translocation
Abstract
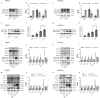
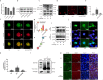
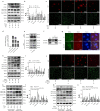
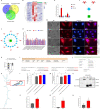
Prevention and treatment of atherosclerosis (AS) by targeting the inflammatory response in vascular endothelial cells has attracted much attention in recent years. Laminar shear stress (LSS) has well-recognized anti-AS properties, however, the exact molecular mechanism remains unclear. In this study, we found that LSS could inhibit the increased expression of intercellular adhesion molecule-1 (ICAM-1), vascular cell adhesion molecule-1 (VCAM-1), cyclooxygenase-2 (COX-2), and matrix metallopeptidase-9 (MMP-9) caused by TNF-α in an autophagy-dependent pathway in human aortic endothelial cells (HAECs) and human umbilical vein endothelial cells (HUVECs). Whole-transcriptome sequencing analysis revealed that erythropoietin-producing hepatocyte receptor B2 (EPHB2) was a key gene in response to LSS. Moreover, co-immunoprecipitation assay indicated that LSS could enhance the EPHB2-mediated nuclear translocation of high mobility group box-1 (HMGB1), which interacts with Beclin-1 (BECN1) and finally leads to autophagy. Simultaneously, we identified an LSS-sensitive long non-coding RNA (lncRNA), LOC10798635, and constructed an LSS-related LOC107986345/miR-128-3p/EPHB2 regulatory axis. Further research revealed the anti-inflammatory effect of LSS depends on autophagy activation resulting from the nuclear translocation of HMGB1 via the LOC107986345/miR-128-3p/EPHB2 axis. Our study demonstrates that LSS could regulate the expression of EPHB2 in HAECs, and the LOC107986345/miR-128-3p/EPHB2 axis plays a vital role in AS development.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Low shear stress induced HMGB1 translocation and release via PECAM-1/PARP-1 pathway to induce inflammation response.PLoS One. 2015 Mar 20;10(3):e0120586. doi: 10.1371/journal.pone.0120586. eCollection 2015. PLoS One. 2015. PMID: 25793984 Free PMC article.
-
Shear-Sensitive lncRNA AF131217.1 Inhibits Inflammation in HUVECs via Regulation of KLF4.Hypertension. 2019 May;73(5):e25-e34. doi: 10.1161/HYPERTENSIONAHA.118.12476. Hypertension. 2019. PMID: 30905197
-
Laminar shear stress alleviates monocyte adhesion and atherosclerosis development via miR-29b-3p/CX3CL1 axis regulation.J Cell Sci. 2022 Jul 15;135(14):jcs259696. doi: 10.1242/jcs.259696. Epub 2022 Jul 22. J Cell Sci. 2022. PMID: 35735031 Free PMC article.
-
Low shear stress-induced blockage of autophagic flux impairs endothelial barrier and facilitates atherosclerosis in mice.Exp Cell Res. 2024 Jun 1;439(1):114071. doi: 10.1016/j.yexcr.2024.114071. Epub 2024 May 8. Exp Cell Res. 2024. PMID: 38729336
-
Berberine inhibits low shear stress-induced vascular endothelial inflammation via decreasing phosphorylation of Akt and IRF3.Tissue Cell. 2022 Dec;79:101946. doi: 10.1016/j.tice.2022.101946. Epub 2022 Sep 23. Tissue Cell. 2022. PMID: 36174269
Cited by
-
The immune system in cardiovascular diseases: from basic mechanisms to therapeutic implications.Signal Transduct Target Ther. 2025 May 23;10(1):166. doi: 10.1038/s41392-025-02220-z. Signal Transduct Target Ther. 2025. PMID: 40404619 Free PMC article. Review.
-
Mechanobiological Approach for Intestinal Mucosal Immunology.Biology (Basel). 2025 Jan 22;14(2):110. doi: 10.3390/biology14020110. Biology (Basel). 2025. PMID: 40001878 Free PMC article. Review.
-
Shear-Sensitive circRNA-LONP2 Promotes Endothelial Inflammation and Atherosclerosis by Targeting NRF2/HO1 Signaling.JACC Basic Transl Sci. 2024 May 27;9(5):652-670. doi: 10.1016/j.jacbts.2024.02.019. eCollection 2024 May. JACC Basic Transl Sci. 2024. PMID: 38984054 Free PMC article.
-
Regulating the regulators: long non-coding RNAs as autophagic controllers in chronic disease management.J Biomed Sci. 2024 Dec 23;31(1):105. doi: 10.1186/s12929-024-01092-9. J Biomed Sci. 2024. PMID: 39716252 Free PMC article. Review.
-
Shear stress induces autophagy in Schlemm's canal cells via primary cilia-mediated SMAD2/3 signaling pathway.Autophagy Rep. 2023;2(1):2236519. doi: 10.1080/27694127.2023.2236519. Epub 2023 Jul 20. Autophagy Rep. 2023. PMID: 37637387 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

