Reversal of the T cell immune system reveals the molecular basis for T cell lineage fate determination in the thymus
- PMID: 35523960
- PMCID: PMC9098387
- DOI: 10.1038/s41590-022-01187-1
Reversal of the T cell immune system reveals the molecular basis for T cell lineage fate determination in the thymus
Abstract
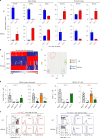
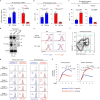
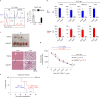
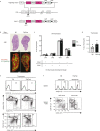
T cell specificity and function are linked during development, as MHC-II-specific TCR signals generate CD4 helper T cells and MHC-I-specific TCR signals generate CD8 cytotoxic T cells, but the basis remains uncertain. We now report that switching coreceptor proteins encoded by Cd4 and Cd8 gene loci functionally reverses the T cell immune system, generating CD4 cytotoxic and CD8 helper T cells. Such functional reversal reveals that coreceptor proteins promote the helper-lineage fate when encoded by Cd4, but promote the cytotoxic-lineage fate when encoded in Cd8-regardless of the coreceptor proteins each locus encodes. Thus, T cell lineage fate is determined by cis-regulatory elements in coreceptor gene loci and is not determined by the coreceptor proteins they encode, invalidating coreceptor signal strength as the basis of lineage fate determination. Moreover, we consider that evolution selected the particular coreceptor proteins that Cd4 and Cd8 gene loci encode to avoid generating functionally reversed T cells because they fail to promote protective immunity against environmental pathogens.
© 2022. This is a U.S. government work and not under copyright protection in the U.S.; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures








Comment in
-
Signal length not strength guides T cell fate.Nat Rev Immunol. 2022 Jul;22(7):407. doi: 10.1038/s41577-022-00736-x. Nat Rev Immunol. 2022. PMID: 35577929 No abstract available.
-
FlipFlop of the T cell immune system.Nat Methods. 2022 Jul;19(7):781. doi: 10.1038/s41592-022-01555-7. Nat Methods. 2022. PMID: 35804242 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

