M6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma
- PMID: 35524319
- PMCID: PMC9074191
- DOI: 10.1186/s12943-022-01575-z
M6A-mediated upregulation of circMDK promotes tumorigenesis and acts as a nanotherapeutic target in hepatocellular carcinoma
Abstract
Background: Emerging evidence suggest the critical role of circular RNAs (circRNAs) in disease development especially in various cancers. However, the oncogenic role of circRNAs in hepatocellular carcinoma (HCC) is still largely unknown.
Methods: RNA sequencing was performed to identify significantly upregulated circRNAs in paired HCC tissues and non-tumor tissues. CCK-8 assay, colony formation, transwell, and xenograft mouse models were used to investigate the role of circRNAs in HCC proliferation and metastasis. Small interfering RNA (siRNA) was used to silence gene expression. RNA immunoprecipitation, biotin pull-down, RNA pull-down, luciferase reporter assay and western blot were used to explore the underlying molecular mechanisms.
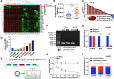
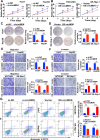
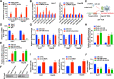
Results: Hsa_circ_0095868, derived from exon 5 of the MDK gene (named circMDK), was identified as a new oncogenic circRNA that was significantly upregulated in HCC. The upregulation of circMDK was associated with the modification of N6-methyladenosine (m6A) and poor survival in HCC patients. Mechanistically, circMDK sponged miR-346 and miR-874-3p to upregulate ATG16L1 (Autophagy Related 16 Like 1), resulting to the activation of PI3K/AKT/mTOR signaling pathway to promote cell proliferation, migration and invasion. Poly (β-amino esters) (PAEs) were synthesized to assist the delivery of circMDK siRNA (PAE-siRNA), which effectively inhibited tumor progression without obvious adverse effects in four liver tumor models including subcutaneous, metastatic, orthotopic and patient-derived xenograft (PDX) models.
Conclusions: CircMDK could serve as a potential tumor biomarker that promotes the progression of HCC via the miR-346/874-3p-ATG16L1 axis. The PAE-based delivery of siRNA improved the stability and efficiency of siRNA targeting circMDK. The PAE-siRNA nanoparticles effectively inhibited HCC proliferation and metastasis in vivo. Our current findings offer a promising nanotherapeutic strategy for the treatment of HCC.
Keywords: ATG16L1; Apoptosis; Hepatocellular carcinoma (HCC); IGF2BP1; N6-methyladenosine (m6A); Nanoparticles (NPs); Poly (β-amino esters) (PAEs); circRNA.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

