Genome-wide analyses of PAM-relaxed Cas9 genome editors reveal substantial off-target effects by ABE8e in rice
- PMID: 35524459
- PMCID: PMC9398351
- DOI: 10.1111/pbi.13838
Genome-wide analyses of PAM-relaxed Cas9 genome editors reveal substantial off-target effects by ABE8e in rice
Abstract
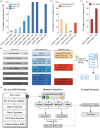
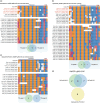
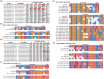
PAM-relaxed Cas9 nucleases, cytosine base editors and adenine base editors are promising tools for precise genome editing in plants. However, their genome-wide off-target effects are largely unexplored. Here, we conduct whole-genome sequencing (WGS) analyses of transgenic plants edited by xCas9, Cas9-NGv1, Cas9-NG, SpRY, nCas9-NG-PmCDA1, nSpRY-PmCDA1 and nSpRY-ABE8e in rice. Our results reveal that Cas9 nuclease and base editors, when coupled with the same guide RNA (gRNA), prefer distinct gRNA-dependent off-target sites. De novo generated gRNAs by SpRY editors lead to additional, but insubstantial, off-target mutations. Strikingly, ABE8e results in ~500 genome-wide A-to-G off-target mutations at TA motif sites per transgenic plant. ABE8e's preference for the TA motif is also observed at the target sites. Finally, we investigate the timeline and mechanism of somaclonal variation due to tissue culture, which chiefly contributes to the background mutations. This study provides a comprehensive understanding on the scale and mechanisms of off-target and background mutations occurring during PAM-relaxed genome editing in plants.
Keywords: PAM-relaxed Cas9 nucleases; adenine base editor; cytosine base editor; genome editing; off-target effect; rice; whole-genome sequencing.
© 2022 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Improving Plant Genome Editing with High-Fidelity xCas9 and Non-canonical PAM-Targeting Cas9-NG.Mol Plant. 2019 Jul 1;12(7):1027-1036. doi: 10.1016/j.molp.2019.03.011. Epub 2019 Mar 27. Mol Plant. 2019. PMID: 30928637
-
PAM-less plant genome editing using a CRISPR-SpRY toolbox.Nat Plants. 2021 Jan;7(1):25-33. doi: 10.1038/s41477-020-00827-4. Epub 2021 Jan 4. Nat Plants. 2021. PMID: 33398158
-
Genome- and transcriptome-wide off-target analyses of a high-efficiency adenine base editor in tomato.Plant Physiol. 2023 Aug 31;193(1):291-303. doi: 10.1093/plphys/kiad347. Plant Physiol. 2023. PMID: 37315207
-
Base editing in rice: current progress, advances, limitations, and future perspectives.Plant Cell Rep. 2021 Apr;40(4):595-604. doi: 10.1007/s00299-020-02656-3. Epub 2021 Jan 10. Plant Cell Rep. 2021. PMID: 33423074 Review.
-
Optimization of genome editing through CRISPR-Cas9 engineering.Bioengineered. 2016 Apr;7(3):166-74. doi: 10.1080/21655979.2016.1189039. Bioengineered. 2016. PMID: 27340770 Free PMC article. Review.
Cited by
-
High-fidelity PAMless base editing of hematopoietic stem cells to treat chronic granulomatous disease.Sci Transl Med. 2024 Oct 16;16(769):eadj6779. doi: 10.1126/scitranslmed.adj6779. Epub 2024 Oct 16. Sci Transl Med. 2024. PMID: 39413163 Free PMC article.
-
Off-target effects in CRISPR/Cas9 gene editing.Front Bioeng Biotechnol. 2023 Mar 9;11:1143157. doi: 10.3389/fbioe.2023.1143157. eCollection 2023. Front Bioeng Biotechnol. 2023. PMID: 36970624 Free PMC article. Review.
-
Efficient plant genome engineering using a probiotic sourced CRISPR-Cas9 system.Nat Commun. 2023 Sep 29;14(1):6102. doi: 10.1038/s41467-023-41802-9. Nat Commun. 2023. PMID: 37773156 Free PMC article.
-
An efficient CRISPR-Cas12a promoter editing system for crop improvement.Nat Plants. 2023 Apr;9(4):588-604. doi: 10.1038/s41477-023-01384-2. Epub 2023 Apr 6. Nat Plants. 2023. PMID: 37024659
-
Versatile plant genome engineering using anti-CRISPR-Cas12a systems.Sci China Life Sci. 2024 Dec;67(12):2730-2745. doi: 10.1007/s11427-024-2704-7. Epub 2024 Aug 15. Sci China Life Sci. 2024. PMID: 39158766
References
-
- Anzalone, A.V. , Koblan, L.W. and Liu, D.R. (2020) Genome editing with CRISPR‐Cas nucleases, base editors, transposases and prime editors. Nat. Biotechnol. 38, 824–844. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

