Refinement of computational identification of somatic copy number alterations using DNA methylation microarrays illustrated in cancers of unknown primary
- PMID: 35524475
- PMCID: PMC9487591
- DOI: 10.1093/bib/bbac161
Refinement of computational identification of somatic copy number alterations using DNA methylation microarrays illustrated in cancers of unknown primary
Abstract
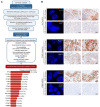
High-throughput genomic technologies are increasingly used in personalized cancer medicine. However, computational tools to maximize the use of scarce tissues combining distinct molecular layers are needed. Here we present a refined strategy, based on the R-package 'conumee', to better predict somatic copy number alterations (SCNA) from deoxyribonucleic acid (DNA) methylation arrays. Our approach, termed hereafter as 'conumee-KCN', improves SCNA prediction by incorporating tumor purity and dynamic thresholding. We trained our algorithm using paired DNA methylation and SNP Array 6.0 data from The Cancer Genome Atlas samples and confirmed its performance in cancer cell lines. Most importantly, the application of our approach in cancers of unknown primary identified amplified potentially actionable targets that were experimentally validated by Fluorescence in situ hybridization and immunostaining, reaching 100% specificity and 93.3% sensitivity.
Keywords: DNA methylation; actionable target identification; cancers of unknown primary; gene amplification; somatic copy number alterations.
© The Author(s) 2022. Published by Oxford University Press.
Figures

References
-
- Berdasco M, Esteller M. Clinical epigenetics: seizing opportunities for translation. Nat Rev Genet 2019;20(2):109–27. - PubMed
-
- Hovestadt V, Zapatka M. Conumee: enhanced copy-number variation analysis using Illumina DNA methylation arrays. R package version 190. http://bioconductor.org/packages/conumee/.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

