Nearest neighbor rules for RNA helix folding thermodynamics: improved end effects
- PMID: 35524574
- PMCID: PMC9122537
- DOI: 10.1093/nar/gkac261
Nearest neighbor rules for RNA helix folding thermodynamics: improved end effects
Abstract
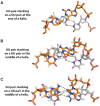
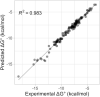
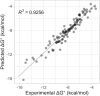
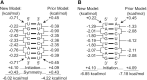
Nearest neighbor parameters for estimating the folding stability of RNA secondary structures are in widespread use. For helices, current parameters penalize terminal AU base pairs relative to terminal GC base pairs. We curated an expanded database of helix stabilities determined by optical melting experiments. Analysis of the updated database shows that terminal penalties depend on the sequence identity of the adjacent penultimate base pair. New nearest neighbor parameters that include this additional sequence dependence accurately predict the measured values of 271 helices in an updated database with a correlation coefficient of 0.982. This refined understanding of helix ends facilitates fitting terms for base pair stacks with GU pairs. Prior parameter sets treated 5'GGUC3' paired to 3'CUGG5' separately from other 5'GU3'/3'UG5' stacks. The improved understanding of helix end stability, however, makes the separate treatment unnecessary. Introduction of the additional terms was tested with three optical melting experiments. The average absolute difference between measured and predicted free energy changes at 37°C for these three duplexes containing terminal adjacent AU and GU pairs improved from 1.38 to 0.27 kcal/mol. This confirms the need for the additional sequence dependence in the model.
© The Author(s) 2022. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
Thermodynamic parameters for an expanded nearest-neighbor model for formation of RNA duplexes with Watson-Crick base pairs.Biochemistry. 1998 Oct 20;37(42):14719-35. doi: 10.1021/bi9809425. Biochemistry. 1998. PMID: 9778347
-
Testing the nearest neighbor model for canonical RNA base pairs: revision of GU parameters.Biochemistry. 2012 Apr 24;51(16):3508-22. doi: 10.1021/bi3002709. Epub 2012 Apr 10. Biochemistry. 2012. PMID: 22490167 Free PMC article.
-
A sensitivity analysis of RNA folding nearest neighbor parameters identifies a subset of free energy parameters with the greatest impact on RNA secondary structure prediction.Nucleic Acids Res. 2017 Jun 2;45(10):6168-6176. doi: 10.1093/nar/gkx170. Nucleic Acids Res. 2017. PMID: 28334976 Free PMC article.
-
Improving RNA nearest neighbor parameters for helices by going beyond the two-state model.Nucleic Acids Res. 2018 Jun 1;46(10):4883-4892. doi: 10.1093/nar/gky270. Nucleic Acids Res. 2018. PMID: 29718397 Free PMC article.
-
The determination of RNA folding nearest neighbor parameters.Methods Mol Biol. 2014;1097:45-70. doi: 10.1007/978-1-62703-709-9_3. Methods Mol Biol. 2014. PMID: 24639154 Review.
Cited by
-
Diversity of Self-Assembled RNA Complexes: From Nanoarchitecture to Nanomachines.Molecules. 2023 Dec 19;29(1):10. doi: 10.3390/molecules29010010. Molecules. 2023. PMID: 38202593 Free PMC article.
-
Analyzing aptamer structure and interactions: in silico modelling and instrumental methods.Biophys Rev. 2024 Nov 20;16(6):685-700. doi: 10.1007/s12551-024-01252-z. eCollection 2024 Dec. Biophys Rev. 2024. PMID: 39830127 Review.
-
Kinetics and dynamics of oligonucleotide hybridization.Nat Rev Chem. 2025 May;9(5):305-327. doi: 10.1038/s41570-025-00704-8. Epub 2025 Apr 11. Nat Rev Chem. 2025. PMID: 40217001 Review.
-
mRNA folding algorithms for structure and codon optimization.Brief Bioinform. 2025 Jul 2;26(4):bbaf386. doi: 10.1093/bib/bbaf386. Brief Bioinform. 2025. PMID: 40755283 Free PMC article. Review.
-
Self-assembly and condensation of intermolecular poly(UG) RNA quadruplexes.Nucleic Acids Res. 2024 Nov 11;52(20):12582-12591. doi: 10.1093/nar/gkae870. Nucleic Acids Res. 2024. PMID: 39373474 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

