Metabotyping the Welsh population of badgers based on thoracic fluid
- PMID: 35524831
- PMCID: PMC9079023
- DOI: 10.1007/s11306-022-01888-6
Metabotyping the Welsh population of badgers based on thoracic fluid
Abstract
Introduction: The European badger (Meles meles) is a known wildlife reservoir for bovine tuberculosis (bTB) and a better understanding of the epidemiology of bTB in this wildlife species is required for disease control in both wild and farmed animals. Flow infusion electrospray-high-resolution mass spectrometry (FIE-HRMS) may potentially identify novel metabolite biomarkers based on which new, rapid, and sensitive point of care tests for bTB infection could be developed.
Objectives: In this foundational study, we engaged on assessing the baseline metabolomic variation in the non-bTB infected badger population ("metabotyping") across Wales.
Methods: FIE-HRMS was applied on thoracic fluid samples obtained by post-mortem of bTB negative badgers (n = 285) which were part of the Welsh Government 'All Wales Badger Found Dead' study.
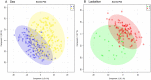
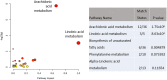
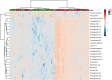
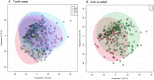
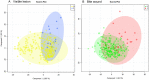
Results: Using principal component analysis and partial least squares-discriminant analyses, the major sources of variation were linked to sex, and to a much lesser extent age, as indicated by tooth wear. Within the female population, variation was seen between lactating and non-lactating individuals. No significant variation linked to the presence of bite wounds, obvious lymphatic lesions or geographical region of origin was observed.
Conclusion: Future metabolomic work when making comparisons between bTB infected and non-infected badger samples will only need be sex-matched and could focus on males only, to avoid lactation bias.
Keywords: All wales badger found dead; Badgers; Diagnostics; High resolution metabolomics; Metabotyping.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Untargeted metabolomic analysis of thoracic blood from badgers indicate changes linked to infection with bovine tuberculosis (Mycobacterium bovis): a pilot study.Metabolomics. 2022 Jul 27;18(8):61. doi: 10.1007/s11306-022-01915-6. Metabolomics. 2022. PMID: 35896834 Free PMC article.
-
Is moving from targeted culling to BCG-vaccination of badgers (Meles meles) associated with an unacceptable increased incidence of cattle herd tuberculosis in the Republic of Ireland? A practical non-inferiority wildlife intervention study in the Republic of Ireland (2011-2017).Prev Vet Med. 2020 Jun;179:105004. doi: 10.1016/j.prevetmed.2020.105004. Epub 2020 Apr 14. Prev Vet Med. 2020. PMID: 32361147
-
Effect of culling and vaccination on bovine tuberculosis infection in a European badger (Meles meles) population by spatial simulation modelling.Prev Vet Med. 2016 Mar 1;125:19-30. doi: 10.1016/j.prevetmed.2015.12.012. Epub 2015 Dec 24. Prev Vet Med. 2016. PMID: 26774448
-
Mycobacterium bovis infection in the Eurasian badger (Meles meles): the disease, pathogenesis, epidemiology and control.J Comp Pathol. 2011 Jan;144(1):1-24. doi: 10.1016/j.jcpa.2010.10.003. Epub 2010 Dec 4. J Comp Pathol. 2011. PMID: 21131004 Review.
-
Wildlife disease ecology from the individual to the population: Insights from a long-term study of a naturally infected European badger population.J Anim Ecol. 2018 Jan;87(1):101-112. doi: 10.1111/1365-2656.12743. Epub 2017 Sep 28. J Anim Ecol. 2018. PMID: 28815647 Review.
References
-
- Abernethy DA, Walton E, Menzies F, Courcier E, Robinson P. Mycobacterium bovis surveillance in European badgers (Meles meles) killed by vehicles in Northern Ireland: An epidemiological evaluation. International Conference on Animal Health Surveillance. 2011;78:216–218.
-
- Broughan JM, Downs SH, Crawshaw TR, Upton PA, Brewer J, Clifton-Hadley RS. Mycobacterium bovis infections in domesticated non-bovine mammalian species. Part 1: Review of epidemiology and laboratory submissions in Great Britain 2004–2010. Veterinary Journal. 2013;198(2):339–345. doi: 10.1016/j.tvjl.2013.09.006. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
