Clinical utility of comprehensive circulating tumor DNA genotyping compared with standard of care tissue testing in patients with newly diagnosed metastatic colorectal cancer
- PMID: 35525184
- PMCID: PMC9271474
- DOI: 10.1016/j.esmoop.2022.100481
Clinical utility of comprehensive circulating tumor DNA genotyping compared with standard of care tissue testing in patients with newly diagnosed metastatic colorectal cancer
Abstract
Background: Comprehensive biomarker testing is essential in selecting optimal treatment for patients with metastatic colorectal cancer (mCRC); however, incomplete genotyping is widespread, with most patients not receiving testing for all guideline-recommended biomarkers, in part due to reliance on burdensome sequential tissue-based single-biomarker tests with long waiting times or availability of only archival tissue samples. We aimed to demonstrate that liquid biopsy, associated with rapid turnaround time (TAT) and lower patient burden, effectively identifies guideline-recommended biomarkers in mCRC relative to standard of care (SOC) tissue testing.
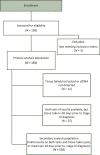
Patients and methods: Prospectively enrolled patients with previously untreated mCRC undergoing physician discretion SOC tissue genotyping submitted pretreatment blood samples for comprehensive circulating tumor DNA (ctDNA) analysis with Guardant360 and targeted RAS and BRAF analysis with OncoBEAM.
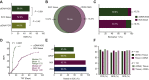
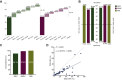
Results: Among 155 patients, physician discretion SOC tissue genotyping identified a guideline-recommended biomarker in 82 patients, versus 88 identified with comprehensive ctDNA (52.9% versus 56.8%, noninferiority demonstrated down to α = 0.005) and 69 identified with targeted PCR ctDNA analysis (52.9% versus 44.5%, noninferiority rejected at α = 0.05). Utilizing ctDNA in addition to tissue increased patient identification for a guideline-recommended biomarker by 19.5% by rescuing those without tissue results either due to tissue insufficiency, test failure, or false negatives. ctDNA median TAT was significantly faster than tissue testing when the complete process from sample acquisition to results was considered (median 10 versus 27 days, P < 0.0001), resulting in accelerated biomarker discovery, with 52.0% biomarker-positive patients identified by ctDNA versus 10.2% by SOC tissue 10 days after sample collection (P < 0.0001).
Conclusions: Comprehensive ctDNA genotyping accurately identifies guideline-recommended biomarkers in patients with mCRC at a rate at least as high as SOC tissue genotyping, in a much shorter time. Based on these findings, the addition of ctDNA genotyping to clinical practice has significant potential to improve the care of patients with mCRC.
Keywords: biomarker; circulating tumor DNA; genomic profiling; liquid biopsy; metastatic colorectal cancer; next-generation sequencing.
Copyright © 2022 The Authors. Published by Elsevier Ltd.. All rights reserved.
Conflict of interest statement
Disclosure MB reports honoraria in an advisory role from Amgen, Bristol Myers Squibb, Merck, MSD, Roche, Sanofi, and Sysmex; received tests for use in study from Guardant Health, Inc. JA-G reports honoraria in a speaker role from Amgen, Bayer, Ipsen, and Roche, and in an advisory role from Merck. SG-C reports honoraria in an advisory role from Servier. MÁ reports honoraria in an advisory role from Bristol Myers Squibb, and in a speaker role from Nanostring, Novartis, and Roche. CR-F reports honoraria in an advisory or speaker role from Sanofi. IA reports honoraria in an advisory role from AstraZeneca, Eli Lilly, Incyte, Ipsen, and Servier. MK, JO, and IF are stockholders and full-time employees at Guardant Health, Inc. EA reports honoraria in an advisory role from AstraZeneca, Daiichi Sankyo, Eli Lilly, Exact Sciences, Novartis, Pfizer, Pierre Fabre, and Roche; has received a research grant from Pfizer. All remaining authors have declared no conflict of interest.
Figures

Similar articles
-
Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non-small Cell Lung Cancer.Clin Cancer Res. 2019 Aug 1;25(15):4691-4700. doi: 10.1158/1078-0432.CCR-19-0624. Epub 2019 Apr 15. Clin Cancer Res. 2019. PMID: 30988079
-
Guardant360 Circulating Tumor DNA Assay Is Concordant with FoundationOne Next-Generation Sequencing in Detecting Actionable Driver Mutations in Anti-EGFR Naive Metastatic Colorectal Cancer.Oncologist. 2020 Mar;25(3):235-243. doi: 10.1634/theoncologist.2019-0441. Epub 2019 Nov 19. Oncologist. 2020. PMID: 32162812 Free PMC article.
-
High concordance of actionable genomic alterations identified between circulating tumor DNA-based and tissue-based next-generation sequencing testing in advanced non-small cell lung cancer: The Korean Lung Liquid Versus Invasive Biopsy Program.Cancer. 2021 Aug 15;127(16):3019-3028. doi: 10.1002/cncr.33571. Epub 2021 Apr 7. Cancer. 2021. PMID: 33826761
-
Clinical Utility of Analyzing Circulating Tumor DNA in Patients with Metastatic Colorectal Cancer.Oncologist. 2018 Nov;23(11):1310-1318. doi: 10.1634/theoncologist.2017-0621. Epub 2018 Apr 26. Oncologist. 2018. PMID: 29700206 Free PMC article. Review.
-
Ongoing Clinical Trials and Future Research Scenarios of Circulating Tumor DNA for the Treatment of Metastatic Colorectal Cancer.Clin Colorectal Cancer. 2024 Dec;23(4):295-308. doi: 10.1016/j.clcc.2024.02.001. Epub 2024 Feb 16. Clin Colorectal Cancer. 2024. PMID: 38519391 Review.
Cited by
-
Clinical Validation of Plasma-Based Genotyping for RAS and BRAF V600E Mutation in Metastatic Colorectal Cancer: SCRUM-Japan GOZILA Substudy.JCO Precis Oncol. 2023 Jun;7:e2200688. doi: 10.1200/PO.22.00688. JCO Precis Oncol. 2023. PMID: 37343204 Free PMC article.
-
Current concepts of anti-EGFR targeting in metastatic colorectal cancer.Front Oncol. 2022 Nov 17;12:1048166. doi: 10.3389/fonc.2022.1048166. eCollection 2022. Front Oncol. 2022. PMID: 36465407 Free PMC article. Review.
-
Rethinking cancer of unknown primary: from diagnostic challenge to targeted treatment.Nat Rev Clin Oncol. 2025 Aug 4. doi: 10.1038/s41571-025-01060-8. Online ahead of print. Nat Rev Clin Oncol. 2025. PMID: 40759731 Review.
-
Private Payer and Medicare Coverage Policies for Use of Circulating Tumor DNA Tests in Cancer Diagnostics and Treatment.J Natl Compr Canc Netw. 2023 Jun;21(6):609-616.e4. doi: 10.6004/jnccn.2023.7011. J Natl Compr Canc Netw. 2023. PMID: 37308126 Free PMC article.
-
Circulating tumor DNA for MRD detection in colorectal cancer: recent advances and clinical implications.Biomark Res. 2025 Jun 23;13(1):89. doi: 10.1186/s40364-025-00796-w. Biomark Res. 2025. PMID: 40551262 Free PMC article. Review.
References
-
- National Comprehensive Cancer Network Colon Cancer (Version 2.2021) http://www.nccn.org/professionals/physician_gls/pdf/colon.pdf Available at.
-
- Mosele F., Remon J., Mateo J., et al. Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2020;31(11):1491–1505. - PubMed
-
- Yoshino T., Arnold D., Taniguchi H., et al. Pan-Asian adapted ESMO consensus guidelines for the management of patients with metastatic colorectal cancer: a JSMO–ESMO initiative endorsed by CSCO, KACO, MOS, SSO and TOS. Ann Oncol. 2018;29(1):44–70. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

