RT-PCR/MALDI-TOF Diagnostic Target Performance Reflects Circulating SARS-CoV-2 Variant Diversity in New York City
- PMID: 35525388
- PMCID: PMC9067105
- DOI: 10.1016/j.jmoldx.2022.04.003
RT-PCR/MALDI-TOF Diagnostic Target Performance Reflects Circulating SARS-CoV-2 Variant Diversity in New York City
Abstract
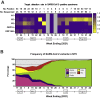
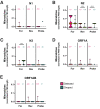
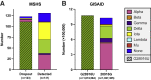
As severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to circulate, multiple variants of concern have emerged. New variants pose challenges for diagnostic platforms because sequence diversity can alter primer/probe-binding sites (PBSs), causing false-negative results. The MassARRAY SARS-CoV-2 Panel (Agena Bioscience) uses RT-PCR and mass spectrometry to detect five multiplex targets across N and ORF1ab genes. Herein, we use a data set of 256 SARS-CoV-2-positive specimens collected between April 11, 2021, and August 28, 2021, to evaluate target performance with paired sequencing data. During this time frame, two targets in the N gene (N2 and N3) were subject to the greatest sequence diversity. In specimens with N3 dropout, 69% harbored the Alpha-specific A28095U polymorphism that introduces a 3'-mismatch to the N3 forward PBS and increases risk of target dropout relative to specimens with 28095A (relative risk, 20.02; 95% CI, 11.36 to 35.72; P < 0.0001). Furthermore, among specimens with N2 dropout, 90% harbored the Delta-specific G28916U polymorphism that creates a 3'-mismatch to the N2 probe PBS and increases target dropout risk (relative risk, 11.92; 95% CI, 8.17 to 14.06; P < 0.0001). These findings highlight the robust capability of MassARRAY SARS-CoV-2 Panel target results to reveal circulating virus diversity, and they underscore the power of multitarget design to capture variants of concern.
Copyright © 2022 Association for Molecular Pathology and American Society for Investigative Pathology. Published by Elsevier Inc. All rights reserved.
Figures

References
-
- Artesi M., Bontems S., Göbbels P., Franckh M., Maes P., Boreux R., Meex C., Melin P., Hayette M.-P., Bours V., Durkin K. A recurrent mutation at position 26340 of SARS-CoV-2 is associated with failure of the E gene quantitative reverse transcription-PCR utilized in a commercial dual-target diagnostic assay. J Clin Microbiol. 2020;58:e01598-20. - PMC - PubMed
-
- Bal A., Destras G., Gaymard A., Stefic K., Marlet J., Eymieux S., Regue H., Semanas Q., d’Aubarede C., Billaud G., Laurent F., Gonzalez C., Mekki Y., Valette M., Bouscambert M., Gaudy-Graffin C., Lina B., Morfin F., Josset L., COVID-Diagnosis HCL Study Group Two-step strategy for the identification of SARS-CoV-2 variant of concern 202012/01 and other variants with spike deletion H69-V70, France, August to December 2020. Euro Surveill. 2021;26:2100008. - PMC - PubMed
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

