Identification of novel key regulatory lncRNAs in gastric adenocarcinoma
- PMID: 35525925
- PMCID: PMC9080188
- DOI: 10.1186/s12864-022-08578-6
Identification of novel key regulatory lncRNAs in gastric adenocarcinoma
Abstract
Background: Stomach adenocarcinoma (STAD) is one of the most common and deadly cancers worldwide. Recent evidence has demonstrated that dysregulation of long noncoding RNAs (lncRNA) is associated with different hallmarks of cancer. lncRNAs also were suggested as novel promising biomarkers for cancer diagnosis and prognosis. Despite these previous investigations, the expression pattern, diagnostic role, and hallmark association of lncRNAs in STAD remain unclear.
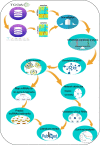
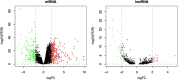
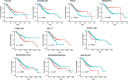
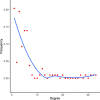
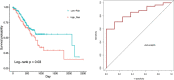
Results: In this study, The STAD lncRNA-mRNA network was constructed based on RNAs that differentially expressed among tumor and normal samples and had a strong expression correlation with others. The high degree nodes of the network were associated with overall survival. In addition, we found that the hubs' regulatory roles have previously been confirmed in different types of cancers by literature. For example, the HCG22 hub inhibited cell proliferation and invasion and induced apoptosis in oral squamous cell carcinoma (OSCC) cells. The levels of PCNA, Vimentin, and Bcl2 were decreased and E-cadherin and Bax expression was elevated in OSCC cells after HCG22 overexpression. Additionally, HCG22 overexpression inhibited the Akt, mTOR, and Wnt/β-catenin pathways. Then lncRNAs were mapped to their related GO terms and cancer hallmarks. Based on these mappings, we predict the hallmarks that might be associated with each lncRNA. Finally, the literature review confirmed our prediction. Among the 20 lncRNAs of the STAD network, 11 lncRNAs (LINC02560, SOX21-AS1, C5orf66-AS1, HCG22, PGM5-AS1, NALT1, ENSG00000241224.2, TINCR, MIR205HG, HNF4A-AS1, ENSG00000262756) demonstrated expression correlation with overall survival (OS). Based on expression analysis, survival analysis, hallmark associations, and literature review, LINC02560, SOX21-AS1, C5orf66-AS1, HCG22, PGM5-AS1, NALT1, ENSG00000241224.2, TINCR, MIR205HG, HNF4A-AS1 plays a regulatory role in STAD. For example, our prediction of association between C5orf66-AS1 expression dysregulation and "sustaining proliferative signal" and "Activating invasion and metastasis" has been confirmed in STAD, OSCC and cervical cancer. Finally, we developed a lncRNA signature with SOX21-AS1 and LINC02560, which classified patients into high and low-risk subgroups with significantly different survival outcomes. The mortality rate of the high-risk patients was significantly higher compared to the low-risk patients (28/1% vs 60.13).
Conclusion: These findings help in designing more precise and detailed experimental studies to find STAD biomarkers and therapeutic targets.
Keywords: Biomarker; Stomach adenocarcinoma; Survival analysis; lncRNA; mRNA.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Screening lncRNAs with diagnostic and prognostic value for human stomach adenocarcinoma based on machine learning and mRNA-lncRNA co-expression network analysis.Mol Genet Genomic Med. 2020 Nov;8(11):e1512. doi: 10.1002/mgg3.1512. Epub 2020 Oct 1. Mol Genet Genomic Med. 2020. PMID: 33002344 Free PMC article.
-
Construction and Validation of a Novel Pyroptosis-Related Four-lncRNA Prognostic Signature Related to Gastric Cancer and Immune Infiltration.Front Immunol. 2022 Mar 22;13:854785. doi: 10.3389/fimmu.2022.854785. eCollection 2022. Front Immunol. 2022. PMID: 35392086 Free PMC article.
-
Angiogenesis-related lncRNAs predict the prognosis signature of stomach adenocarcinoma.BMC Cancer. 2021 Dec 7;21(1):1312. doi: 10.1186/s12885-021-08987-y. BMC Cancer. 2021. PMID: 34876056 Free PMC article.
-
The role of long non-coding RNA AFAP1-AS1 in human malignant tumors.Pathol Res Pract. 2018 Oct;214(10):1524-1531. doi: 10.1016/j.prp.2018.08.014. Epub 2018 Aug 20. Pathol Res Pract. 2018. PMID: 30173945 Review.
-
Identification of MFI2-AS1, a Novel Pivotal lncRNA for Prognosis of Stage III/IV Colorectal Cancer.Dig Dis Sci. 2020 Dec;65(12):3538-3550. doi: 10.1007/s10620-020-06064-1. Epub 2020 Jan 20. Dig Dis Sci. 2020. PMID: 31960204 Review.
Cited by
-
The role of MAPK pathway in gastric cancer: unveiling molecular crosstalk and therapeutic prospects.J Transl Med. 2024 Dec 24;22(1):1142. doi: 10.1186/s12967-024-05998-8. J Transl Med. 2024. PMID: 39719645 Free PMC article. Review.
-
Construction and validation of an anoikis-related long non-coding RNA-based prognostic model for head and neck squamous cell carcinoma.Transl Cancer Res. 2025 Jul 30;14(7):4160-4178. doi: 10.21037/tcr-2024-2520. Epub 2025 Jul 25. Transl Cancer Res. 2025. PMID: 40792140 Free PMC article.
-
Intelligent structure prediction and visualization analysis of non-coding RNA in osteosarcoma research.Front Oncol. 2024 Mar 12;14:1255061. doi: 10.3389/fonc.2024.1255061. eCollection 2024. Front Oncol. 2024. PMID: 38532928 Free PMC article.
-
Crosslinking B-Cell Lymphoma (BCL1) in Surgery Patients by Exploring Its Therapeutic Potential for Head and Neck Cancer Pathology.Cureus. 2025 Apr 23;17(4):e82853. doi: 10.7759/cureus.82853. eCollection 2025 Apr. Cureus. 2025. PMID: 40416279 Free PMC article.
-
Unraveling the influence of LncRNA in gastric cancer pathogenesis: a comprehensive review focus on signaling pathways interplay.Med Oncol. 2024 Aug 5;41(9):218. doi: 10.1007/s12032-024-02455-w. Med Oncol. 2024. PMID: 39103705 Review.
References
-
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG, Lagarde J, Veeravalli L, Ruan X, Ruan Y, Lassmann T, Carninci P, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012;22(9):1775–1789. doi: 10.1101/gr.132159.111. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

