Chromosomal instability as a source of genomic plasticity
- PMID: 35526333
- PMCID: PMC9156567
- DOI: 10.1016/j.gde.2022.101913
Chromosomal instability as a source of genomic plasticity
Abstract
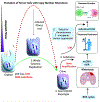
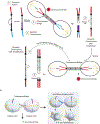
Chromosomal instability (CIN) is a hallmark of the most aggressive malignancies. Features of these tumors include complex genomic rearrangements, the presence of mis-segregated chromosomes in micronuclei, and extrachromosomal DNA (ecDNA) formation. Here, we review the development of CIN, and examine CIN in the context of cancer evolution, tumor genomic evolution, and therapeutic resistance. We also discuss the role of whole-genome duplications, breakage-fusion-bridge cycles, ecDNA or double minutes in gene amplification promoting tumor evolution.
Keywords: Cancer Evolution; Chromosomal Instability; Micronuclei; ecDNA.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Figures

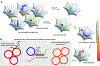
References
-
- Hanahan D, Weinberg RA: Hallmarks of cancer: the next generation. Cell 2011, 144:646–674. - PubMed
-
- Bartek J, Bartkova J, Lukas J: DNA damage signalling guards against activated oncogenes and tumour progression. Oncogene 2007, 26:7773–7779. - PubMed
-
- Lukas C, Falck J, Bartkova J, Bartek J, Lukas J: Distinct spatiotemporal dynamics of mammalian checkpoint regulators induced by DNA damage. Nat Cell Biol 2003, 5:255–260. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

