Neutralization of SARS-CoV-2 Omicron sub-lineages BA.1, BA.1.1, and BA.2
- PMID: 35526534
- PMCID: PMC9035359
- DOI: 10.1016/j.chom.2022.04.014
Neutralization of SARS-CoV-2 Omicron sub-lineages BA.1, BA.1.1, and BA.2
Abstract
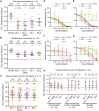
Recent reports of SARS-CoV-2 Omicron variant sub-lineages, BA.1, BA.1.1, and BA.2, have reignited concern over potential escape from vaccine- and infection-induced immunity. We examine the sensitivity of these sub-lineages and other major variants to neutralizing antibodies from mRNA-vaccinated and boosted individuals, as well as recovered COVID-19 patients, including those infected with Omicron. We find that all Omicron sub-lineages, especially BA.1 and BA.1.1, exhibit substantial immune escape that is largely overcome by mRNA vaccine booster doses. While Omicron BA.1.1 escapes almost completely from neutralization by early-pandemic COVID-19 patient sera and to a lesser extent from sera of Delta-infected patients, BA.1.1 is sensitive to Omicron-infected patient sera. Critically, all Omicron sub-lineages, including BA.2, are comparably neutralized by Omicron patient sera. These results highlight the importance of booster vaccine doses for protection against all Omicron variants and provide insight into the immunity from natural infection against Omicron sub-lineages.
Keywords: BA.1; BA.1.1; BA.2; COVID-19; Omicron; SARS-CoV-2; mRNA vaccine; neutralizing antibody.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures


References
-
- Abbas Q., Kusakin A., Sharrouf K., Jyakhwo S., Komissarov A.S. Follow-up investigation and detailed mutational characterization of the SARS-CoV-2 Omicron variant lineages (BA. 1, BA. 2, BA. 3 and BA. 1.1) bioRxiv. 2022 doi: 10.1101/2022.02.25.481941. Preprint at. - DOI
-
- Bednarski E., Del Rio Estrada P.M., DaSilva J., Boukadida C., Zhang F., Villalobos Y.A.L., Range X.R., Isidro E.P., Garcia E.L., Rivera D.D., et al. Antibody and memory B-cell immunity in a heterogeneously SARS-CoV-2 infected and vaccinated population. medRxiv. 2022 doi: 10.1101/2022.02.07.22270626. Preprint at. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

