Genomics of Ancient Pathogens: First Advances and Prospects
- PMID: 35526849
- PMCID: PMC8916790
- DOI: 10.1134/S0006297922030051
Genomics of Ancient Pathogens: First Advances and Prospects
Abstract
Paleogenomics is one of the urgent and promising areas of interdisciplinary research in the today's world science. New genomic methods of ancient DNA (aDNA) analysis, such as next generation sequencing (NGS) technologies, make it possible not only to obtain detailed genetic information about historical and prehistoric human populations, but also to study individual microbial and viral pathogens and microbiomes from different ancient and historical objects. Studies of aDNA of pathogens by reconstructing their genomes have so far yielded complete sequences of the ancient pathogens that played significant role in the history of the world: Yersinia pestis (plague), Variola virus (smallpox), Vibrio cholerae (cholera), HBV (hepatitis B virus), as well as the equally important endemic human infectious agents: Mycobacterium tuberculosis (tuberculosis), Mycobacterium leprae (leprosy), and Treponema pallidum (syphilis). Genomic data from these pathogens complemented the information previously obtained by paleopathologists and allowed not only to identify pathogens from the past pandemics, but also to recognize the pathogen lineages that are now extinct, to refine chronology of the pathogen appearance in human populations, and to reconstruct evolutionary history of the pathogens that are still relevant to public health today. In this review, we describe state-of-the-art genomic research of the origins and evolution of many ancient pathogens and viruses and examine mechanisms of the emergence and spread of the ancient infections in the mankind history.
Keywords: ancient DNA; cholera; human populations; leprosy; paleogenomics; paleopathology; pathogen; plague; smallpox; syphilis; tuberculosis.
Conflict of interest statement
Authors declare no conflicts of interests. This article does not contain description of studies with the involvement of humans or animal subjects.
Figures

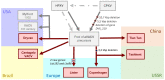
References
-
- Buikstra, J. E., and Roberts, C. (2012) The Global History of Paleopathology: Pioneers and Prospects, Oxford Univ. Press.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

