A novel ENU-induced Cpox mutation causes microcytic hypochromic anemia in mice
- PMID: 35527013
- PMCID: PMC9671764
- DOI: 10.1538/expanim.22-0032
A novel ENU-induced Cpox mutation causes microcytic hypochromic anemia in mice
Abstract
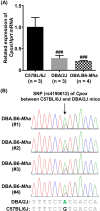
Mouse models of red blood cell abnormalities are important for understanding the underlying molecular mechanisms of human erythrocytic diseases. DBA.B6-Mha (Microcytic hypochromic anemia) congenic mice were generated from the cross between N-ethyl-N-nitrosourea (ENU)-mutagenized male C57BL/6J and female DBA/2J mice as part of the RIKEN large-scale ENU mutagenesis project. The mice were established by backcrossing with DBA/2J mice for more than 20 generations. These mice showed autosomal-dominant microcytic hypochromic anemia with decreased mean corpuscular volume (MCV) and mean corpuscular hemoglobin (MCH) levels and increased red blood cell distribution width (RDW) and plasma ferritin levels. Linkage analysis indicated that the Mha locus was located within an interval of approximately 1.95-Mb between D16Nut1 (58.35 Mb) and D16Mit185 (60.30 Mb) on mouse chromosome 16. Mutation analysis revealed that DBA.B6-Mha mice had a point mutation (c.921-2A>G) at the acceptor site of intron 4 in the coproporphyrinogen oxidase (Cpox) gene, a heme-synthesizing gene. RT-PCR revealed that the Cpox mRNA in DBA.B6-Mha mice caused splicing errors. Our results suggest that microcytic hypochromic anemia in DBA.B6-Mha mice is owing to impaired heme synthesis caused by splice mutations in Cpox. Therefore, the DBA.B6-Mha mice may be used to elucidate the molecular mechanisms underlying microcytic hypochromic anemia caused by mutations in Cpox. Although low MCV levels are known to confer malarial resistance to the host, there were no marked changes in the susceptibility of DBA.B6-Mha mice to rodent malarial (Plasmodium yoelii 17XL) infection.
Keywords: N-ethyl-N-nitrosourea (ENU) mutagenesis; coproporphyrinogen oxidase (Cpox) gene; malaria; mice; microcytic hypochromic anemia.
Figures


Similar articles
-
A mouse model of hereditary coproporphyria identified in an ENU mutagenesis screen.Dis Model Mech. 2017 Aug 1;10(8):1005-1013. doi: 10.1242/dmm.029116. Epub 2017 Jun 9. Dis Model Mech. 2017. PMID: 28600349 Free PMC article.
-
Cystathionine-γ-lyase (CSE) deficiency increases erythropoiesis and promotes mitochondrial electron transport via the upregulation of coproporphyrinogen III oxidase and consequent stimulation of heme biosynthesis.Biochem Pharmacol. 2019 Nov;169:113604. doi: 10.1016/j.bcp.2019.08.006. Epub 2019 Aug 14. Biochem Pharmacol. 2019. PMID: 31421132 Free PMC article.
-
hem6: an ENU-induced recessive hypochromic microcytic anemia mutation in the mouse.Blood. 2008 Nov 15;112(10):4308-13. doi: 10.1182/blood-2007-09-111500. Epub 2008 Sep 9. Blood. 2008. PMID: 18780836 Free PMC article.
-
alpha-globin gene deletion and point mutation analysis among in Iranian patients with microcytic hypochromic anemia.Haematologica. 2003 Oct;88(10):1196-7. Haematologica. 2003. PMID: 14555321 Review.
-
Molecular basis of inherited microcytic anemia due to defects in iron acquisition or heme synthesis.Haematologica. 2009 Mar;94(3):395-408. doi: 10.3324/haematol.13619. Epub 2009 Jan 30. Haematologica. 2009. PMID: 19181781 Free PMC article. Review.
Cited by
-
PHEXL222P Mutation Increases Phex Expression in a New ENU Mouse Model for XLH Disease.Genes (Basel). 2022 Jul 28;13(8):1356. doi: 10.3390/genes13081356. Genes (Basel). 2022. PMID: 36011266 Free PMC article.
-
BALB.NCT-Cpox nct is a unique mouse model of hereditary coproporphyria.Mol Genet Metab Rep. 2023 Mar 16;35:100964. doi: 10.1016/j.ymgmr.2023.100964. eCollection 2023 Jun. Mol Genet Metab Rep. 2023. PMID: 36967721 Free PMC article.
References
-
- Hughes MR, Anderson N, Maltby S, Wong J, Berberovic Z, Birkenmeier CS, et al. A novel ENU-generated truncation mutation lacking the spectrin-binding and C-terminal regulatory domains of Ank1 models severe hemolytic hereditary spherocytosis. Exp Hematol. 2011; 39: 305–320, 320.e1–320.e2. doi: 10.1016/j.exphem.2010.12.009 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

