Understanding the R882H mutation effects of DNA methyltransferase DNMT3A: a combination of molecular dynamics simulations and QM/MM calculations
- PMID: 35527972
- PMCID: PMC9072302
- DOI: 10.1039/c9ra06791d
Understanding the R882H mutation effects of DNA methyltransferase DNMT3A: a combination of molecular dynamics simulations and QM/MM calculations
Abstract
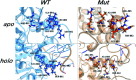
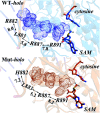
DNA (cytosine-5)-methyltransferase 3A (DNMT3A), a key enzyme for de novo epigenetic methylation in human beings, was reported to undergo an R882H mutation in approximately 25% of M4/M5 subtype acute myeloid leukemia (AML) patients. In this work, a combination of classical molecular dynamics (MD) simulations and QM/MM calculation methods was utilized to reveal the molecular mechanism behind the activity attenuation caused by R882H mutation. We found that R882H mutation induces a "folded" conformation in the methyl donor S-adenosylmethionine (SAM) through different types of hydrogen bond formation at the terminal carbonyl oxygen atom and the hydroxyl O3' atom of the ribose ring on SAM, with Arg891 as a mediator. Energetically, both the pre-reaction state (PRS) and transition state (TS) were stabilized in the R882H mutant. However, the energy barrier of the rate-determining step from the PRS to the TS was calculated to be roughly 1.0 kcal mol-1 larger in the R882H mutant than the WT. Also, a dynamic transformation occurred along the helix where R882H was located, tending to manifest in a quasi-"Newton's cradle" manner from the mutational site to the active site residues of DNMT3A. Our computational results provided molecular insights into the pathogenic R882H mutation and advanced the understanding of its mechanism.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





Similar articles
-
Preferential Self-interaction of DNA Methyltransferase DNMT3A Subunits Containing the R882H Cancer Mutation Leads to Dominant Changes of Flanking Sequence Preferences.J Mol Biol. 2022 Apr 15;434(7):167482. doi: 10.1016/j.jmb.2022.167482. Epub 2022 Feb 5. J Mol Biol. 2022. PMID: 35131259
-
The R882H DNMT3A hot spot mutation stabilizes the formation of large DNMT3A oligomers with low DNA methyltransferase activity.J Biol Chem. 2019 Nov 8;294(45):16966-16977. doi: 10.1074/jbc.RA119.010126. Epub 2019 Oct 3. J Biol Chem. 2019. PMID: 31582562 Free PMC article.
-
DNMT3A R882H mutation drives daunorubicin resistance in acute myeloid leukemia via regulating NRF2/NQO1 pathway.Cell Commun Signal. 2022 Oct 27;20(1):168. doi: 10.1186/s12964-022-00978-1. Cell Commun Signal. 2022. PMID: 36303144 Free PMC article.
-
AML-Associated Mutations in DNA Methyltransferase DNMT3A.Biochemistry (Mosc). 2021 Mar;86(3):307-318. doi: 10.1134/S000629792103007X. Biochemistry (Mosc). 2021. PMID: 33838631 Review.
-
Acute myeloid leukemia-associated DNMT3A p.Arg882His mutation in a patient with Tatton-Brown-Rahman overgrowth syndrome as a constitutional mutation.Am J Med Genet A. 2017 Jan;173(1):250-253. doi: 10.1002/ajmg.a.37995. Epub 2016 Nov 7. Am J Med Genet A. 2017. PMID: 27991732 Review.
Cited by
-
Unveiling the methyl transfer mechanisms in the epigenetic machinery DNMT3A-3 L: A comprehensive study integrating assembly dynamics with catalytic reactions.Comput Struct Biotechnol J. 2023 Mar 5;21:2086-2099. doi: 10.1016/j.csbj.2023.03.002. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 36968013 Free PMC article.
-
Evolutionary coupling-inspired engineering of alcohol dehydrogenase reveals the influence of distant sites on its catalytic efficiency for stereospecific synthesis of chiral alcohols.Comput Struct Biotechnol J. 2021 Oct 26;19:5864-5873. doi: 10.1016/j.csbj.2021.10.031. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34815831 Free PMC article.
-
Insights into the Inhibitory Mechanisms of the Covalent Drugs for DNMT3A.Int J Mol Sci. 2023 Aug 10;24(16):12652. doi: 10.3390/ijms241612652. Int J Mol Sci. 2023. PMID: 37628829 Free PMC article.

