Exploring the chemical space and the bioactivity profile of lactams: a chemoinformatic study
- PMID: 35528563
- PMCID: PMC9070607
- DOI: 10.1039/c9ra04841c
Exploring the chemical space and the bioactivity profile of lactams: a chemoinformatic study
Abstract
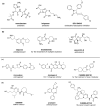
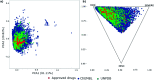
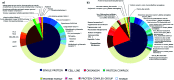
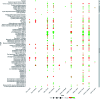
Lactams are a class of compounds important for drug design, due to their great variety of potential therapeutic applications, spanning cancer, diabetes, and infectious diseases. So far, the biological profile and chemical diversity of lactams have not been characterized in a systematic and detailed manner. In this work, we report the chemoinformatic analysis of beta-, gamma-, delta- and epsilon-lactams present in databases of approved drugs, natural products, and bioactive compounds from the large public database ChEMBL. We identified the main biological targets in which the lactams have been evaluated according to their chemical classification. We also identified the most frequent scaffolds and those that can be prioritized in chemical synthesis, since they are scaffolds with potential biological activity but with few reported analogs. Results of the biological and chemoinformatic analysis of lactams indicate that spiro- and bridged-lactams belong to classes with the lowest number of compounds and unique scaffolds, and some showing activity against specific targets. Information obtained from this analysis allows focusing the design of new chemical structures in less explored spaces and with increased possibilities of success.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare that they do not have any conflict of interest related to this manuscript.
Figures




References
LinkOut - more resources
Full Text Sources

