A model for the chemomechanical coupling of myosin-V molecular motors
- PMID: 35528596
- PMCID: PMC9070430
- DOI: 10.1039/c9ra05072h
A model for the chemomechanical coupling of myosin-V molecular motors
Abstract
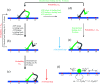
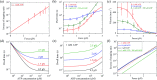
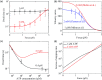
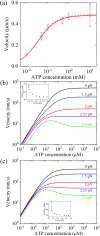
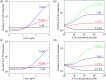
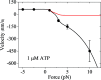
Herein, a model for the chemomechanical coupling of dimeric myosin-V motors is presented. Based on this model and the proposal that the rate constants of the ATPase activity of the two heads are independent of an external force in a range smaller than the stall force, we analytically studied the dynamics of the motor, such as the stepping ratio, dwell time between two mechanical steps, and velocity, under varying force and ATP concentrations. The theoretical results well reproduce the diverse available single-molecule experimental data. In particular, the experimental data showing that at a low ATP concentration, the dwell time and velocity have less force dependency than at a high ATP concentration is explained quantitatively. Moreover, the dependency of the chemomechanical coupling ratio on the force and ATP concentration was studied.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
No conflict of interest exists.
Figures

References
LinkOut - more resources
Full Text Sources

