Optimized Transformation and Gene Editing of the B104 Public Maize Inbred by Improved Tissue Culture and Use of Morphogenic Regulators
- PMID: 35528934
- PMCID: PMC9072829
- DOI: 10.3389/fpls.2022.883847
Optimized Transformation and Gene Editing of the B104 Public Maize Inbred by Improved Tissue Culture and Use of Morphogenic Regulators
Abstract
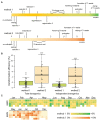
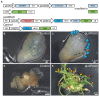
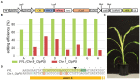
Plant transformation is a bottleneck for the application of gene editing in plants. In Zea mays (maize), a breakthrough was made using co-transformation of the morphogenic transcription factors BABY BOOM (BBM) and WUSCHEL (WUS) to induce somatic embryogenesis. Together with adapted tissue culture media, this was shown to increase transformation efficiency significantly. However, use of the method has not been reported widely, despite a clear need for increased transformation capacity in academic settings. Here, we explore use of the method for the public maize inbred B104 that is widely used for transformation by the research community. We find that only modifying tissue culture media already boosts transformation efficiency significantly and can reduce the time in tissue culture by 1 month. On average, production of independent transgenic plants per starting embryo increased from 1 to 4% using BIALAPHOS RESISTANCE (BAR) as a selection marker. In addition, we reconstructed the BBM-WUS morphogenic gene cassette and evaluated its functionality in B104. Expression of the morphogenic genes under tissue- and development stage-specific promoters led to direct somatic embryo formation on the scutellum of zygotic embryos. However, eight out of ten resulting transgenic plants showed pleiotropic developmental defects and were not fertile. This undesirable phenotype was positively correlated with the copy number of the morphogenic gene cassette. Use of constructs in which morphogenic genes are flanked by a developmentally controlled Cre/LoxP recombination system led to reduced T-DNA copy number and fertile T0 plants, while increasing transformation efficiency from 1 to 5% using HIGHLY-RESISTANT ACETOLACTATE SYNTHASE as a selection marker. Addition of a CRISPR/Cas9 module confirmed functionality for gene editing applications, as exemplified by editing the gene VIRESCENT YELLOW-LIKE (VYL) that can act as a visual marker for gene editing in maize. The constructs, methods, and insights produced in this work will be valuable to translate the use of BBM-WUS and other emerging morphogenic regulators (MRs) to other genotypes and crops.
Keywords: Agrobacterium; CRISPR/Cas9; T-DNA; gene editing; maize; morphogenic genes; tissue culture; transformation.
Copyright © 2022 Aesaert, Impens, Coussens, Van Lerberge, Vanderhaeghen, Desmet, Vanhevel, Bossuyt, Wambua, Van Lijsebettens, Inzé, De Keyser, Jacobs, Karimi and Pauwels.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Akoyi J., Mgutu A. J., Machuka J., Van Lijsebettens M., Taracha C., Anami S. E. (2013). Dicamba growth regulator promotes genotype independent somatic embryogenesis from immature zygotic embryos of tropical maize inbred lines. J. Life Sci. 7, 677–689. doi: 10.17265/1934-7391/2013.07.001 - DOI
-
- Aliu E., Azanu M. K., Wang K., Lee K. (2020). Generation of thymidine auxotrophic Agrobacterium tumefaciens strains for plant transformation. bioRxiv [Preprint]. doi: 10.1101/2020.08.21.261941 - DOI
-
- Anand A., Arling M. L., da Silva Conceicao A., Gordon-Kamm W. J., Hastings C. E., Hoerster G. M., et al. . (2016). 615 Methods and Compositions for Rapid Plant Transformation. US20170121722A1.
LinkOut - more resources
Full Text Sources
Other Literature Sources

