WHIRLIES Are Multifunctional DNA-Binding Proteins With Impact on Plant Development and Stress Resistance
- PMID: 35528945
- PMCID: PMC9070903
- DOI: 10.3389/fpls.2022.880423
WHIRLIES Are Multifunctional DNA-Binding Proteins With Impact on Plant Development and Stress Resistance
Abstract
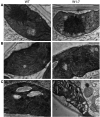
WHIRLIES are plant-specific proteins binding to DNA in plastids, mitochondria, and nucleus. They have been identified as significant components of nucleoids in the organelles where they regulate the structure of the nucleoids and diverse DNA-associated processes. WHIRLIES also fulfil roles in the nucleus by interacting with telomers and various transcription factors, among them members of the WRKY family. While most plants have two WHIRLY proteins, additional WHIRLY proteins evolved by gene duplication in some dicot families. All WHIRLY proteins share a conserved WHIRLY domain responsible for ssDNA binding. Structural analyses revealed that WHIRLY proteins form tetramers and higher-order complexes upon binding to DNA. An outstanding feature is the parallel localization of WHIRLY proteins in two or three cell compartments. Because they translocate from organelles to the nucleus, WHIRLY proteins are excellent candidates for transducing signals between organelles and nucleus to allow for coordinated activities of the different genomes. Developmental cues and environmental factors control the expression of WHIRLY genes. Mutants and plants with a reduced abundance of WHIRLY proteins gave insight into their multiple functionalities. In chloroplasts, a reduction of the WHIRLY level leads to changes in replication, transcription, RNA processing, and DNA repair. Furthermore, chloroplast development, ribosome formation, and photosynthesis are impaired in monocots. In mitochondria, a low level of WHIRLIES coincides with a reduced number of cristae and a low rate of respiration. The WHIRLY proteins are involved in the plants' resistance toward abiotic and biotic stress. Plants with low levels of WHIRLIES show reduced responsiveness toward diverse environmental factors, such as light and drought. Consequently, because such plants are impaired in acclimation, they accumulate reactive oxygen species under stress conditions. In contrast, several plant species overexpressing WHIRLIES were shown to have a higher resistance toward stress and pathogen attacks. By their multiple interactions with organelle proteins and nuclear transcription factors maybe a comma can be inserted here? and their participation in organelle-nucleus communication, WHIRLY proteins are proposed to serve plant development and stress resistance by coordinating processes at different levels. It is proposed that the multifunctionality of WHIRLY proteins is linked to the plasticity of land plants that develop and function in a continuously changing environment.
Keywords: DNA-binding; WHIRLY; development; nucleoid; stress.
Copyright © 2022 Krupinska, Desel, Frank and Hensel.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Artsatbanov V. Y., Vostroknutova G. N., Shleeva M. O., Goncharenko A. V., Zinin A. I., Ostrovsky D. N., et al. (2012). Influence of oxidative and nitrosative stress on accumulation of diphosphate intermediates of the non-mevalonate pathway of isoprenoid biosynthesis in Corynebacteria and mycobacteria. Biochemistry 77, 362–371. doi: 10.1134/S0006297912040074, PMID: - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources

