Prognostic Role and Potential Mechanisms of N6-methyladenosine-related Long Noncoding RNAs in Hepatocellular Carcinoma
- PMID: 35528973
- PMCID: PMC9039697
- DOI: 10.14218/JCTH.2021.00096
Prognostic Role and Potential Mechanisms of N6-methyladenosine-related Long Noncoding RNAs in Hepatocellular Carcinoma
Abstract
Background and aims: Numerous studies have explored the important role of N6-methyladenosine (m6A) in cancer. Nonetheless, the interaction between m6A and long noncoding RNAs (lncRNAs) is poorly investigated. Herein, we systematically analyzed the role and prognostic value of m6A-related lncRNAs in hepatocellular carcinoma (HCC).
Methods: The m6A-related lncRNAs were identified based on the correlation coefficients with m6A-related genes in HCC from The Cancer Genome Atlas. Subsequently, a novel risk score model was determined using the least absolute shrinkage and selection operator Cox regression analyses. Univariate and multivariate Cox analyses were used to identify independent prognostic factors for overall survival (OS) of HCC; thereafter, a prognostic nomogram was constructed.
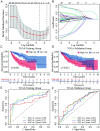
Results: A total of 259 lncRNAs showed significant correlations with m6A in HCC, while 29 lncRNAs had prognostic significance. Further, six critical m6A-related lncRNAs (NRAV, SNHG3, KDM4A-AS1, AC074117.1, AC025176.1, and AL031985.3) were screened out to construct a novel risk score model which classified HCC patients into high- and low-risk groups. Survival analyses revealed that patients in the high-risk group exhibited worse OS, both in the training and validation groups. The risk score was also identified as an independent prognostic factor of OS, and a nomogram was established and verified with superior prediction capacity. Besides, the risk score significantly correlated with the expression of immune checkpoint genes and immune subtypes.
Conclusions: These findings indicated the significant role of m6A-related lncRNAs in HCC and the potential application of the novel risk score model for prognostic prediction.
Keywords: Hepatocellular carcinoma; Immune checkpoints; Long noncoding RNAs; N6-methyladenosine; Prognosis.
© 2022 Authors.
Conflict of interest statement
The authors have no conflict of interests related to this publication.
Figures






References
LinkOut - more resources
Full Text Sources
