Enhanced extracellular recombinant keratinase activity in Bacillus subtilis SCK6 through signal peptide optimization and site-directed mutagenesis
- PMID: 35529123
- PMCID: PMC9073338
- DOI: 10.1039/c9ra07866e
Enhanced extracellular recombinant keratinase activity in Bacillus subtilis SCK6 through signal peptide optimization and site-directed mutagenesis
Abstract
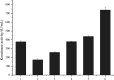
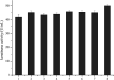
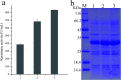
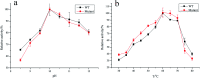
Keratinase has a great commercial value owing to its applications in the enzymatic dehairing of goatskins. In this study, we adopted a combined strategy to enhance the extracellular recombinant keratinase activity in Bacillus subtilis SCK6. First, nine signal peptides were screened to enhance the expression of extracellular keratinase. The recombinant strain with SPLipA exhibited the highest extracellular keratinase activity of 739.03 U per mL, which was two-fold higher activity of the wild type. Second, based on the multiple sequence alignment with the bacterial alkaline proteases, the mutant (M123L/V149I/A242N) was introduced into the keratinase. Comparing with the wild type of keratinase, the mutant M123L/V149I/A242N showed an increase in the extracellular keratinase activity, which was about 1.2-fold higher activity of the wild type. Finally, the keratinase expression vector with SPLipA and mutant M123L/V149I/A242N was constructed, and the extracellular keratinase activity reported at 830.91 U per mL was a 2.2-fold activity of the wild type. Then, the mutant keratinase was purified and characterized. The mutant exhibited properties similar to those of the wild type at an optimal temperature of 60 °C and pH 10.0. Conclusively, the extracellular expression of keratinase was enhanced via a combined strategy, and the mutant keratinase demonstrated properties similar to that of the wild type of keratinase.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
No potential conflict of interest was reported by the authors.
Figures



Similar articles
-
High-expression keratinase by Bacillus subtilis SCK6 for enzymatic dehairing of goatskins.Int J Biol Macromol. 2019 Aug 15;135:119-126. doi: 10.1016/j.ijbiomac.2019.05.131. Epub 2019 May 21. Int J Biol Macromol. 2019. PMID: 31125653
-
Construction of a Rapid Feather-Degrading Bacterium by Overexpression of a Highly Efficient Alkaline Keratinase in Its Parent Strain Bacillus amyloliquefaciens K11.J Agric Food Chem. 2016 Jan 13;64(1):78-84. doi: 10.1021/acs.jafc.5b04747. Epub 2015 Dec 24. J Agric Food Chem. 2016. PMID: 26671753
-
Cloning and expression of keratinase gene in Bacillus megaterium and optimization of fermentation conditions for the production of keratinase by recombinant strain.J Appl Microbiol. 2007 Oct;103(4):1301-10. doi: 10.1111/j.1365-2672.2007.03372.x. J Appl Microbiol. 2007. PMID: 17897234
-
A novel alkaline keratinase from Bacillus subtilis DP1 with potential utility in cosmetic formulation.Int J Biol Macromol. 2016 Jun;87:256-62. doi: 10.1016/j.ijbiomac.2016.02.067. Epub 2016 Mar 3. Int J Biol Macromol. 2016. PMID: 26940376
-
Molecular strategies to increase keratinase production in heterologous expression systems for industrial applications.Appl Microbiol Biotechnol. 2021 May;105(10):3955-3969. doi: 10.1007/s00253-021-11321-y. Epub 2021 May 3. Appl Microbiol Biotechnol. 2021. PMID: 33937928 Review.
Cited by
-
The flexible linker and CotG were more effective for the spore surface display of keratinase KERQ7.World J Microbiol Biotechnol. 2023 Dec 7;40(1):35. doi: 10.1007/s11274-023-03854-3. World J Microbiol Biotechnol. 2023. PMID: 38057620
-
Keratinases as Versatile Enzymatic Tools for Sustainable Development.Biomolecules. 2021 Dec 18;11(12):1900. doi: 10.3390/biom11121900. Biomolecules. 2021. PMID: 34944542 Free PMC article. Review.
-
Heterologous Expression of Recombinant Transglutaminase in Bacillus subtilis SCK6 with Optimized Signal Peptide and Codon, and Its Impact on Gelatin Properties.J Microbiol Biotechnol. 2020 Jul 28;30(7):1082-1091. doi: 10.4014/jmb.2002.02049. J Microbiol Biotechnol. 2020. PMID: 32325545 Free PMC article.
-
High keratinase and other types of hydrolase activity of the new strain of Bacillus paralicheniformis.PLoS One. 2024 Oct 25;19(10):e0312679. doi: 10.1371/journal.pone.0312679. eCollection 2024. PLoS One. 2024. PMID: 39453952 Free PMC article.
-
New Bacillus paralicheniformis strain with high proteolytic and keratinolytic activity.Sci Rep. 2024 Sep 30;14(1):22621. doi: 10.1038/s41598-024-73468-8. Sci Rep. 2024. PMID: 39349615 Free PMC article.
References
-
- Paul T. Jana A. Mandal A. K. Mandal A. Das Mohpatra P. K. Mondal K. C. Sustainable Chem. Pharm. 2016;3:8–22. doi: 10.1016/j.scp.2016.01.001. - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources

