Role of Long Noncoding RNAs in Smoking-Induced Lung Cancer: An In Silico Study
- PMID: 35529255
- PMCID: PMC9070410
- DOI: 10.1155/2022/7169353
Role of Long Noncoding RNAs in Smoking-Induced Lung Cancer: An In Silico Study
Abstract
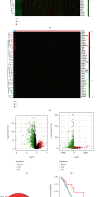
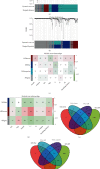
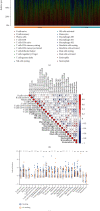
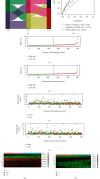
The prevalence of lung cancer induced by cigarette smoking has increased over time. Long noncoding (lnc) RNAs, regulatory factors that play a role in human diseases, are commonly dysregulated in lung cancer. Cigarette smoking is closely related to changes in lncRNA expression, which can affect lung cancer. Herein, we assess the mechanism of lung cancer initiation induced by smoking. To calculate the impact of smoking on the survival of patients with lung cancer, we extracted data from The Cancer Genome Atlas and Gene Expression Omnibus databases and identified the differentially expressed genes in the lung cancer tissue compared to the normal lung tissue. Genes positively and negatively associated with smoking were identified. Gene Ontology, Kyoto Encyclopedia of Genes and Genomes, and Cytoscape analyses were performed to determine the function of the genes and the effects of smoking on the immune microenvironment. lncRNAs corresponding to smoking-associated genes were identified, and a smoking-related lncRNA model was constructed using univariate and multivariate Cox analyses. This model was used to assess the survival of and potential risk in patients who smoked. During screening, 562 differentially expressed genes were identified, and we elucidated that smoking affected the survival of patients 4.5 years after the diagnosis of lung cancer. Furthermore, genes negatively associated with smoking were closely associated with immunity. Twelve immune cell types were also found to infiltrate differentially in smokers and nonsmokers. Thus, the smoking-associated lncRNA model is a good predictor of survival and risk in smokers and may be used as an independent prognostic factor for lung cancer.
Copyright © 2022 Lei Ge et al.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures



References
-
- Allemani C., Matsuda T., Di Carlo V., et al. Global surveillance of trends in cancer survival 2000-14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet . 2018;391(10125):1023–1075. doi: 10.1016/S0140-6736(17)33326-3. - DOI - PMC - PubMed
-
- National Cancer Institute. SEER stat fact sheets: lung and bronchus cancer. 2018, http://seer.cancer.gov/statfacts/html/lungb.html.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

