Integrated lipidomics and RNA sequencing analysis reveal novel changes during 3T3-L1 cell adipogenesis
- PMID: 35529487
- PMCID: PMC9074861
- DOI: 10.7717/peerj.13417
Integrated lipidomics and RNA sequencing analysis reveal novel changes during 3T3-L1 cell adipogenesis
Abstract
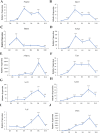
After adipogenic differentiation, key regulators of adipogenesis are stimulated and cells begin to accumulate lipids. To identify specific changes in lipid composition and gene expression patterns during 3T3-L1 cell adipogenesis, we carried out lipidomics and RNA sequencing analysis of undifferentiated and differentiated 3T3-L1 cells. The analysis revealed significant changes in lipid content and gene expression patterns during adipogenesis. Slc2a4 was up-regulated, which may enhance glucose transport; Gpat3, Agpat2, Lipin1 and Dgat were also up-regulated, potentially to enrich intracellular triacylglycerol (TG). Increased expression levels of Pnpla2, Lipe, Acsl1 and Lpl likely increase intracellular free fatty acids, which can then be used for subsequent synthesis of other lipids, such as sphingomyelin (SM) and ceramide (Cer). Enriched intracellular diacylglycerol (DG) can also provide more raw materials for the synthesis of phosphatidylinositol (PI), phosphatidylcholine (PC), phosphatidylethanolamine (PE), ether-PE, and ether-PC, whereas high expression of Pla3 may enhance the formation of lysophophatidylcholine (LPC) and lysophosphatidylethanolamine (LPE). Therefore, in the process of adipogenesis of 3T3-L1 cells, a series of genes are activated, resulting in large changes in the contents of various lipid metabolites in the cells, especially TG, DG, SM, Cer, PI, PC, PE, etherPE, etherPC, LPC and LPE. These findings provide a theoretical basis for our understanding the pathophysiology of obesity.
Keywords: 3T3-L1; Adipogenesis; Ceramide; Diacylglycerol; Lipidomics; Phospholipid; Sphingomyelin; Triacylglycerol.
©2022 Pei et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures









References
-
- Al Adhami H, Evano B, Le Digarcher A, Gueydan C, Dubois E, Parrinello H, Dantec C, Bouschet T, Varrault A, Journot L. A systems-level approach to parental genomic imprinting: the imprinted gene network includes extracellular matrix genes and regulates cell cycle exit and differentiation. Genome Research. 2015;25:353–367. doi: 10.1101/gr.175919.114. - DOI - PMC - PubMed
-
- Bremer J, Figard PH, Greenberg DM. The biosynthesis of choline and its relation to phospholipid metabolism. Biochimica et Biophysica Acta. 1960;43:477–488. doi: 10.1016/0006-3002(60)90470-4. - DOI
-
- Cao J, Li JL, Li D, Tobin JF, Gimeno RE. Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:19695–19700. doi: 10.1073/pnas.0609140103. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

