miR-9-5p promotes myogenic differentiation via the Dlx3/Myf5 axis
- PMID: 35529491
- PMCID: PMC9074878
- DOI: 10.7717/peerj.13360
miR-9-5p promotes myogenic differentiation via the Dlx3/Myf5 axis
Abstract
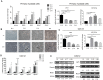
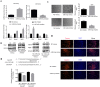
MicroRNAs play an important role in myogenic differentiation, they bind to target genes and regulate muscle formation. We previously found that miR-9-5p, which is related to bone formation, was increased over time during the process of myogenic differentiation. However, the mechanism by which miR-9-5p regulates myogenic differentiation remains largely unknown. In the present study, we first examined myotube formation and miR-9-5p, myogenesis-related genes including Dlx3, Myod1, Mef2c, Desmin, MyoG and Myf5 expression under myogenic induction. Then, we detected the expression of myogenic transcription factors after overexpression or knockdown of miR-9-5p or Dlx3 in the mouse premyoblast cell line C2C12 by qPCR, western blot and myotube formation under myogenic induction. A luciferase assay was performed to confirm the regulatory relationships between not only miR-9-5p and Dlx3 but also Dlx3 and its downstream gene, Myf5, which is an essential transcription factor of myogenic differentiation. The results showed that miR-9-5p promoted myogenic differentiation by increasing myogenic transcription factor expression and promoting myotube formation, but Dlx3 exerted the opposite effect. Moreover, the luciferase assay showed that miR-9-5p bound to the 3'UTR of Dlx3 and downregulated Dlx3 expression. Dlx3 in turn suppressed Myf5 expression by binding to the Myf5 promoter, ultimately inhibiting the process of myogenic differentiation. In conclusion, the miR-9-5p/Dlx3/Myf5 axis is a novel pathway for the regulation of myogenic differentiation, and can be a potential target to treat the diseases related to muscle dysfunction.
Keywords: Dlx3; MiR-9-5p; Myf5; Myogenic differentiation.
©2022 Dong et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures



Similar articles
-
MiR-183-5p induced by saturated fatty acids regulates the myogenic differentiation by directly targeting FHL1 in C2C12 myoblasts.BMB Rep. 2020 Nov;53(11):605-610. doi: 10.5483/BMBRep.2020.53.11.175. BMB Rep. 2020. PMID: 33148375 Free PMC article.
-
miR-10b-5p Regulates C2C12 Myoblasts Proliferation and Differentiation.Biosci Biotechnol Biochem. 2019 Feb;83(2):291-299. doi: 10.1080/09168451.2018.1533805. Epub 2018 Oct 18. Biosci Biotechnol Biochem. 2019. PMID: 30336746
-
MiR-96-5p Induced by Palmitic Acid Suppresses the Myogenic Differentiation of C2C12 Myoblasts by Targeting FHL1.Int J Mol Sci. 2020 Dec 11;21(24):9445. doi: 10.3390/ijms21249445. Int J Mol Sci. 2020. PMID: 33322515 Free PMC article.
-
Distal-less homeobox 3, a negative regulator of myogenesis, is downregulated by microRNA-133.J Cell Biochem. 2019 Feb;120(2):2226-2235. doi: 10.1002/jcb.27533. Epub 2018 Sep 11. J Cell Biochem. 2019. PMID: 30277585
-
Function of the myogenic regulatory factors Myf5, MyoD, Myogenin and MRF4 in skeletal muscle, satellite cells and regenerative myogenesis.Semin Cell Dev Biol. 2017 Dec;72:19-32. doi: 10.1016/j.semcdb.2017.11.011. Epub 2017 Nov 15. Semin Cell Dev Biol. 2017. PMID: 29127046 Review.
Cited by
-
Genome-wide characteristics and potential functions of circular RNAs from the embryo muscle development in Chengkou mountain chicken.Front Vet Sci. 2024 May 30;11:1375042. doi: 10.3389/fvets.2024.1375042. eCollection 2024. Front Vet Sci. 2024. PMID: 38872802 Free PMC article.
-
Fetal Brain-Derived Exosomal miRNAs from Maternal Blood: Potential Diagnostic Biomarkers for Fetal Alcohol Spectrum Disorders (FASDs).Int J Mol Sci. 2024 May 27;25(11):5826. doi: 10.3390/ijms25115826. Int J Mol Sci. 2024. PMID: 38892014 Free PMC article.
References
-
- Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Tréguer K, Carmona G, Bonauer A, Horrevoets AJ, Didier N, Girmatsion Z, Biliczki P, Ehrlich JR, Katus HA, Müller OJ, Potente M, Zeiher AM, Hermeking H, Dimmeler S. MicroRNA-34a regulates cardiac ageing and function. Nature. 2013;495:107–110. doi: 10.1038/nature11919. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

