Network subgraph-based approach for analyzing and comparing molecular networks
- PMID: 35529499
- PMCID: PMC9074881
- DOI: 10.7717/peerj.13137
Network subgraph-based approach for analyzing and comparing molecular networks
Abstract
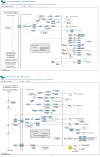
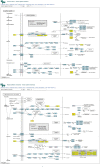
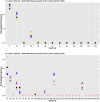
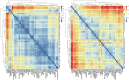
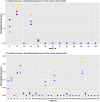
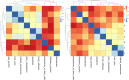
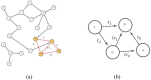
Molecular networks are built up from genetic elements that exhibit feedback interactions. Here, we studied the problem of measuring the similarity of directed networks by proposing a novel alignment-free approach: the network subgraph-based approach. Our approach does not make use of randomized networks to determine modular patterns embedded in a network, and this method differs from the network motif and graphlet methods. Network similarity was quantified by gauging the difference between the subgraph frequency distributions of two networks using Jensen-Shannon entropy. We applied the subgraph approach to study three types of molecular networks, i.e., cancer networks, signal transduction networks, and cellular process networks, which exhibit diverse molecular functions. We compared the performance of our subgraph detection algorithm with other algorithms, and the results were consistent, but other algorithms could not address the issue of subgraphs/motifs embedded within a subgraph/motif. To evaluate the effectiveness of the subgraph-based method, we applied the method along with the Jensen-Shannon entropy to classify six network models, and it achieves a 100% accuracy of classification. The proposed information-theoretic approach allows us to determine the structural similarity of two networks regardless of node identity and network size. We demonstrated the effectiveness of the subgraph approach to cluster molecular networks that exhibit similar regulatory interaction topologies. As an illustration, our method can identify (i) common subgraph-mediated signal transduction and/or cellular processes in AML and pancreatic cancer, and (ii) scaffold proteins in gastric cancer and hepatocellular carcinoma; thus, the results suggested that there are common regulation modules for cancer formation. We also found that the underlying substructures of the molecular networks are dominated by irreducible subgraphs; this feature is valid for the three classes of molecular networks we studied. The subgraph-based approach provides a systematic scenario for analyzing, compare and classifying molecular networks with diverse functionalities.
Keywords: Cancer networks; Cellular processes; Entropy; Graph theory; Information theory; Jensen-Shannon entropy; Network subgraphs; Signal transduction networks.
©2022 Huang et al.
Conflict of interest statement
The authors declare there are no competing interests.
Figures




References
-
- Aparìcio DO, Ribeiro PMP, Silva FMA. Network comparison using directed graphlets. 2015. 1511.01964
-
- Bagrow JP, Bollt EM. An information-theoretic, all-scales approach to comparing networks. Applied Network Science. 2019;4:1–15. doi: 10.1007/s41109-018-0108-x. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

