Enhanced stability of an intrinsically disordered protein against proteolytic cleavage through interactions with silver nanoparticles
- PMID: 35529627
- PMCID: PMC9071183
- DOI: 10.1039/c9ra05514b
Enhanced stability of an intrinsically disordered protein against proteolytic cleavage through interactions with silver nanoparticles
Abstract
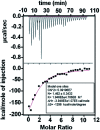
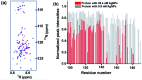
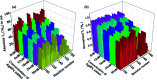
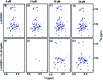
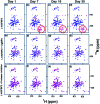
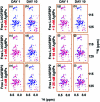
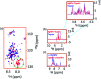
Intrinsically disordered proteins (IDPs), being sensitive to proteolytic degradation both in vitro and in vivo, can be stabilized by the interactions with various binding partners. Here, we show for the first time that silver nanoparticles (AgNPs) have the ability to enhance the half-life of an IDP, thereby rendering it stable for a month against proteolytic degradation. The conjugate of the unstructured linker domain of human insulin-like growth factor binding protein-2 (L-hIGFBP2) with 10 nm citrate-capped AgNPs was studied using two-dimensional NMR spectroscopy and other biophysical techniques. Our studies reveal the extent and nature of residue-specific interactions of the IDP with AgNPs. These interactions mask proteolysis-prone sites of the IDP and stabilize it. This study opens new avenues for the design of appropriate nanoparticles targeting IDPs and for storage, stabilization and delivery of IDPs into cells in a stable form.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures




References
LinkOut - more resources
Full Text Sources

