Systematic analysis of ferroptosis-related long non-coding RNA predicting prognosis in patients with lung squamous cell carcinoma
- PMID: 35529787
- PMCID: PMC9073741
- DOI: 10.21037/tlcr-22-224
Systematic analysis of ferroptosis-related long non-coding RNA predicting prognosis in patients with lung squamous cell carcinoma
Abstract
Background: Ferroptosis is a novel iron-dependent cell death, and an increasing number of studies have shown that long non-coding RNA (lncRNAs) are involved in the ferroptosis process. However, studies on ferroptosis-related lncRNAs in lung squamous cell carcinoma (LUSC) are limited. In addition, the prognostic role of ferroptosis-related lncRNAs and their relationship with the immune microenvironment and methylation of LUSC is unclear. This study aimed to investigate the potential prognostic value of ferroptosis-related lncRNAs and their involved biological functions in LUSC.
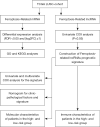
Methods: The Cancer Genome Atlas (TCGA) database and the FerrDb website were used to obtain ferroptosis-related genes for LUSC. The "limma" R package and Pearson analysis were used to find ferroptosis-related lncRNAs. The biological functions of the characterized lncRNAs were analyzed by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). We evaluated the prognostic power of this model using Kaplan-Meier analysis, receiver operating characteristic (ROC), and decision curve analysis (DCA). Univariate and multifactor Cox (proportional-hazards) risk model and a nomogram were produced using risk models and clinicopathological parameters for further verification. In addition, the relationship between characterized lncRNAs and tumor immune infiltration and methylation was also discussed.
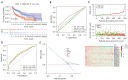
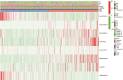
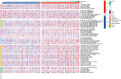
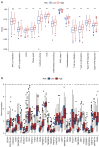
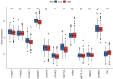
Results: We identified 29 characterized lncRNAs to produce prognostic risk models. Kaplan-Meier analysis revealed the high-risk group was associated with poor prognosis in LUSC (P<0.001), and ROC (AUC =0.658) and DCA suggested that risk models could predict prognosis. Univariate and multifactorial Cox as well as nomogram further validated the prognostic model (P<0.001). Gene set enrichment analysis (GSEA) showed that the high-risk group was associated with pro-tumor pathways and high-frequency mutations in TP53 were present in both groups. Single sample gene set enrichment analysis (ssGSEA) showed significant differences in immune cell infiltration subtypes and corresponding functions between the two groups. Some immune checkpoint and methylation-related genes were significantly different between the two groups (P<0.05).
Conclusions: We investigated the potential mechanisms of LUSC development from the perspective of ferroptosis-related lncRNAs, providing new insights into LUSC research, and identified 29 lncRNAs as biomarkers to predict the prognosis of LUSC patients.
Keywords: Ferroptosis; long non-coding RNA (lncRNA); lung squamous cell carcinoma (LUSC); prognostic signature.
2022 Translational Lung Cancer Research. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-22-224/coif). Sujie Ni serves as an unpaid editorial board member of Translational Lung Cancer Research from June 2017 to June 2022. The other authors have no conflicts of interest to declare.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
