The generation of PD-L1 and PD-L2 in cancer cells: From nuclear chromatin reorganization to extracellular presentation
- PMID: 35530130
- PMCID: PMC9069407
- DOI: 10.1016/j.apsb.2021.09.010
The generation of PD-L1 and PD-L2 in cancer cells: From nuclear chromatin reorganization to extracellular presentation
Abstract
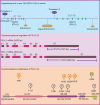
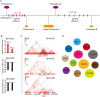
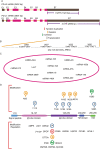
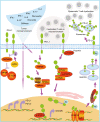
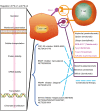
The immune checkpoint blockade (ICB) targeting on PD-1/PD-L1 has shown remarkable promise in treating cancers. However, the low response rate and frequently observed severe side effects limit its broad benefits. It is partially due to less understanding of the biological regulation of PD-L1. Here, we systematically and comprehensively summarized the regulation of PD-L1 from nuclear chromatin reorganization to extracellular presentation. In PD-L1 and PD-L2 highly expressed cancer cells, a new TAD (topologically associating domain) (chr9: 5,400,000-5,600,000) around CD274 and CD273 was discovered, which includes a reported super-enhancer to drive synchronous transcription of PD-L1 and PD-L2. The re-shaped TAD allows transcription factors such as STAT3 and IRF1 recruit to PD-L1 locus in order to guide the expression of PD-L1. After transcription, the PD-L1 is tightly regulated by miRNAs and RNA-binding proteins via the long 3'UTR. At translational level, PD-L1 protein and its membrane presentation are tightly regulated by post-translational modification such as glycosylation and ubiquitination. In addition, PD-L1 can be secreted via exosome to systematically inhibit immune response. Therefore, fully dissecting the regulation of PD-L1/PD-L2 and thoroughly detecting PD-L1/PD-L2 as well as their regulatory networks will bring more insights in ICB and ICB-based combinational therapy.
Keywords: 3′-UTR, 3′-untranslated region; ADAM17, a disintegrin and metalloprotease 17; APCs, antigen-presenting cells; AREs, adenylate and uridylate (AU)-rich elements; ATF3, activating transcription factor 3; CD273/274, cluster of differentiation 273/274; CDK4, cyclin-dependent kinase 4; CMTM6, CKLF like MARVEL transmembrane domain containing 6; CSN5, COP9 signalosome subunit 5; CTLs, cytotoxic T lymphocytes; EMT, epithelial to mesenchymal transition; EpCAM, epithelial cell adhesion molecule; Exosome; FACS, fluorescence-activated cell sorting; GSDMC, Gasdermin C; GSK3β, glycogen synthase kinase 3 beta; HSF1, heat shock transcription factor 1; Hi-C, high throughput chromosome conformation capture; ICB, immune checkpoint blockade; IFN, interferon; IL-6, interleukin 6; IRF1, interferon regulatory factor 1; Immune checkpoint blockade; JAK, Janus kinase 1; NFκB, nuclear factor kappa B; NSCLC, non-small cell lung cancer; OTUB1, OTU deubiquitinase, ubiquitin aldehyde binding 1; PARP1, poly(ADP-ribose) polymerase 1; PD-1, programmed cell death-1; PD-L1; PD-L1, programmed death-ligand 1; PD-L2; PD-L2, programmed death ligand 2; Post-transcriptional regulation; Post-translational regulation; SP1, specificity protein 1; SPOP, speckle-type POZ protein; STAG2, stromal antigen 2; STAT3, signal transducer and activator of transcription 3; T2D, type 2 diabetes; TADs, topologically associating domains; TFEB, transcription factor EB; TFs, transcription factors; TNFα, tumor necrosis factor-alpha; TTP, tristetraprolin; Topologically associating domain; Transcription; UCHL1, ubiquitin carboxy-terminal hydrolase L1; USP22, ubiquitin specific peptidase 22; dMMR, deficient DNA mismatch repair; irAEs, immune related adverse events.
© 2022 Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences. Production and hosting by Elsevier B.V.
Figures
References
-
- Postow M.A., Sidlow R., Hellmann M.D. Immune-related adverse events associated with immune checkpoint blockade. N Engl J Med. 2018;378:158–168. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

