Inhibition of the CDK9-cyclin T1 protein-protein interaction as a new approach against triple-negative breast cancer
- PMID: 35530158
- PMCID: PMC9069406
- DOI: 10.1016/j.apsb.2021.10.024
Inhibition of the CDK9-cyclin T1 protein-protein interaction as a new approach against triple-negative breast cancer
Abstract
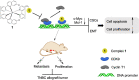
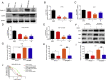
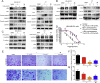
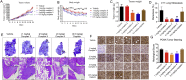
Cyclin-dependent kinase 9 (CDK9) activity is correlated with worse outcomes of triple-negative breast cancer (TNBC) patients. The heterodimer between CDK9 with cyclin T1 is essential for maintaining the active state of the kinase and targeting this protein-protein interaction (PPI) may offer promising avenues for selective CDK9 inhibition. Herein, we designed and generated a library of metal complexes bearing the 7-chloro-2-phenylquinoline CˆN ligand and tested their activity against the CDK9-cyclin T1 PPI. Complex 1 bound to CDK9 via an enthalpically-driven binding mode, leading to disruption of the CDK9-cyclin T1 interaction in vitro and in cellulo. Importantly, complex 1 showed promising anti-metastatic activity against TNBC allografts in mice and was comparably active compared to cisplatin. To our knowledge, 1 is the first CDK9-cyclin T1 PPI inhibitor with anti-metastatic activity against TNBC. Complex 1 could serve as a new platform for the future design of more efficacious kinase inhibitors against cancer, including TNBC.
Keywords: Epigenetics; Kinase inhibitor; Metal complex; Metastasis; Protein–protein interaction; Triple-negative breast cancer.
© 2022 Chinese Pharmaceutical Association and Institute of Materia Medica, Chinese Academy of Medical Sciences. Production and hosting by Elsevier B.V.
Figures



References
-
- Cheng S.S., Yang G.J., Wang W.H., Ma D.L., Leung C.H. Discovery of a tetrahydroisoquinoline-based CDK9‒cyclin T1 protein–protein interaction inhibitor as an anti-proliferative and anti-migration agent against triple-negative breast cancer cells. Genes Dis. 2021 https://www.sciencedirect.com/science/article/pii/S2352304221000866 Available from: - PMC - PubMed
-
- Yang G.J., Wang W., Mok S.W.F., Wu C., Law B.Y.K., Miao X.M., et al. Selective inhibition of lysine-specific demethylase 5A (KDM5A) using a rhodium (III) complex for triple-negative breast cancer therapy. Angew Chem Int Ed. 2018;57:13091–13095. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

