ZAP70 Activation Compensates for Loss of Class IA PI3K Isoforms Through Activation of the JAK-STAT3 Pathway
- PMID: 35530641
- PMCID: PMC9066532
- DOI: 10.21873/cdp.10122
ZAP70 Activation Compensates for Loss of Class IA PI3K Isoforms Through Activation of the JAK-STAT3 Pathway
Abstract
Background/aim: Tyrosine kinases have crucial functions in cell signaling and proliferation. The phosphatidylinositol 3-kinase (PI3K) pathway is frequently deregulated in human cancer and is an essential regulator of cellular proliferation. We aimed to determine which tyrosine kinases contribute to resistance elicited by PI3K silencing and inhibition.
Materials and methods: To mimic catalytic inactivation of p110α/β, specific p110α (BYL719) and p110β (KIN193) inhibitors were used in addition to genetic knock-out in in vitro assays. Cell viability was assessed using crystal violet staining, whereas cellular transformation ability was analyzed by soft-agar growth assays.
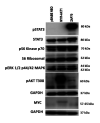
Results: Activated zeta chain of T-cell receptor-associated protein kinase 70 (ZAP70) generated resistance to PI3K inhibition. This resistance was via activation of the Janus kinase/signal transducer and activator of transcription 3 (JAK/STAT3) axis. We demonstrated that activated ZAP70 has a high transforming capability associated with the formation of malignant phenotype in untransformed cells and has the potential to be a tumor-initiating factor in cancer cells.
Conclusion: ZAP70 may be a potent driver of proliferation and transformation in untransformed cells and is implicated in resistance to PI3K inhibitors in cancer cells.
Keywords: PI3K; ZAP70; cell signaling; growth resistance; tyrosine kinases.
Copyright 2022, International Institute of Anticancer Research.
Conflict of interest statement
The Authors declare no competing conflicts of interest.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
