Circulating microRNAs in seminal plasma as predictors of sperm retrieval in microdissection testicular sperm extraction
- PMID: 35530943
- PMCID: PMC9073789
- DOI: 10.21037/atm-21-5100
Circulating microRNAs in seminal plasma as predictors of sperm retrieval in microdissection testicular sperm extraction
Abstract
Background: Because of focal spermatogenesis in some nonobstructive azoospermia (NOA) patients, testicular spermatozoa can be retrieved by microdissection testicular sperm extraction (micro-TESE) for intracytoplasmic sperm injection (ICSI) to achieve successful fertilization. Currently, testicular biopsy is widely performed for the prognosis of micro-TESE; however, it might miss foci with active spermatogenesis because of the 'blind manner' of puncture, highlighting the needs for biomarkers that could indicate actual spermatogenesis conditions in the testis. Thus, we screened microRNAs in the seminal plasma for potential biomarkers to provide a non-invasive and reliable preoperative assessment for micro-TESE.
Methods: We screened the seminal plasma microRNAs from NOA patients with and without sperm retrieval (n=6 in each group) together with fertile men (n=6) by RNA sequencing, and the selected microRNAs were validated by quantitative polymerase chain reaction (qPCR). Next, a predictive model was established by performing ordered logistic regression using the qPCR data of 56 specimens, and the predictive accuracy of this model was evaluated using 40 more specimens in a blind manner.
Results: Four microRNAs (hsa-miR-34b-3p, hsa-miR-34c-3p, hsa-miR-3065-3p, and hsa-miR-4446-3p) were identified as biomarkers, and the predictive model Logit = 2.0881+ 0.13448 mir-34b-3p + 0.58679 mir-34c-3p + 0.15636 mir-3065-3p + 0.09523 mir-4446-3p was established by machine learning. The model provided a high predictive accuracy (AUC =0.927).
Conclusions: We developed a predictive model with high accuracy for micro-TESE, with which NOA patients might obtain accurate assessment of spermatogenesis conditions in testes before surgery.
Keywords: Nonobstructive azoospermia; biomarker; microRNA; microdissection testicular sperm extraction (micro-TESE); spermatogenesis.
2022 Annals of Translational Medicine. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-5100/coif). The authors have no conflicts of interest to declare.
Figures

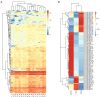
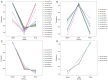


Similar articles
-
Potential of testis-derived circular RNAs in seminal plasma to predict the outcome of microdissection testicular sperm extraction in patients with idiopathic non-obstructive azoospermia.Hum Reprod. 2021 Sep 18;36(10):2649-2660. doi: 10.1093/humrep/deab196. Hum Reprod. 2021. PMID: 34477868
-
Circulatory exosomal tRF-Glu-CTC-005 and tRF-Gly-GCC-002 serve as predictive factors of successful microdissection testicular sperm extraction in patients with nonobstructive azoospermia.Fertil Steril. 2022 Mar;117(3):512-521. doi: 10.1016/j.fertnstert.2021.11.010. Epub 2021 Dec 24. Fertil Steril. 2022. PMID: 34955241
-
MicroRNA profile comparison of testicular tissues derived from successful and unsuccessful microdissection testicular sperm extraction retrieval in non-obstructive azoospermia patients.Reprod Fertil Dev. 2019 Apr;31(4):671-682. doi: 10.1071/RD17423. Reprod Fertil Dev. 2019. PMID: 30423284
-
Microdissection testicular sperm extraction (micro-TESE) in men with infertility due to nonobstructive azoospermia: summary of current literature.Int Urol Nephrol. 2021 Nov;53(11):2193-2210. doi: 10.1007/s11255-021-02979-4. Epub 2021 Aug 19. Int Urol Nephrol. 2021. PMID: 34410586 Review.
-
Testis sperm extraction.Asian J Urol. 2015 Apr;2(2):79-84. doi: 10.1016/j.ajur.2015.04.018. Epub 2015 Apr 16. Asian J Urol. 2015. PMID: 29264124 Free PMC article. Review.
Cited by
-
An artificial neural network model to diagnose non-obstructive azoospermia based on RNA-binding protein-related genes.Aging (Albany NY). 2023 Apr 24;15(8):3120-3140. doi: 10.18632/aging.204674. Epub 2023 Apr 24. Aging (Albany NY). 2023. PMID: 37116198 Free PMC article.
-
A comparison of the expression patterns and diagnostic capability of the ncRNAs NEAT1 and miR-34a in non-obstructive azoospermia and severe oligospermia.Hum Genomics. 2025 Mar 31;19(1):35. doi: 10.1186/s40246-025-00742-9. Hum Genomics. 2025. PMID: 40165339 Free PMC article.
-
Predictors of Successful Testicular Sperm Extraction: A New Era for Men with Non-Obstructive Azoospermia.Biomedicines. 2024 Nov 25;12(12):2679. doi: 10.3390/biomedicines12122679. Biomedicines. 2024. PMID: 39767586 Free PMC article. Review.
-
Human sperm RNA in male infertility.Nat Rev Urol. 2025 Feb;22(2):92-115. doi: 10.1038/s41585-024-00920-9. Epub 2024 Sep 10. Nat Rev Urol. 2025. PMID: 39256514 Review.
-
Seminal plasma biomarkers for predicting successful sperm retrieval in patients with nonobstructive azoospermia: a narrative review of human studies.Basic Clin Androl. 2023 Apr 20;33(1):9. doi: 10.1186/s12610-023-00184-0. Basic Clin Androl. 2023. PMID: 37076787 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
