Potentials, prospects and applications of genome editing technologies in livestock production
- PMID: 35531207
- PMCID: PMC9072931
- DOI: 10.1016/j.sjbs.2021.11.037
Potentials, prospects and applications of genome editing technologies in livestock production
Abstract
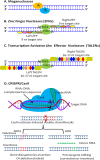
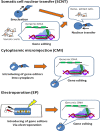
In recent years, significant progress has been achieved in genome editing applications using new programmable DNA nucleases such as zinc finger nucleases (ZFNs), transcription activator-like endonucleases (TALENs) and the clustered regularly interspaced short palindromic repeats/Cas9 system (CRISPR/Cas9). These genome editing tools are capable of nicking DNA precisely by targeting specific sequences, and enable the addition, removal or substitution of nucleotides via double-stranded breakage at specific genomic loci. CRISPR/Cas system, one of the most recent genome editing tools, affords the ability to efficiently generate multiple genomic nicks in single experiment. Moreover, CRISPR/Cas systems are relatively easy and cost effective when compared to other genome editing technologies. This is in part because CRISPR/Cas systems rely on RNA-DNA binding, unlike other genome editing tools that rely on protein-DNA interactions, which affords CRISPR/Cas systems higher flexibility and more fidelity. Genome editing tools have significantly contributed to different aspects of livestock production such as disease resistance, improved performance, alterations of milk composition, animal welfare and biomedicine. However, despite these contributions and future potential, genome editing technologies also have inherent risks, and therefore, ethics and social acceptance are crucial factors associated with implementation of these technologies. This review emphasizes the impact of genome editing technologies in development of livestock breeding and production in numerous species such as cattle, pigs, sheep and goats. This review also discusses the mechanisms behind genome editing technologies, their potential applications, risks and associated ethics that should be considered in the context of livestock.
Keywords: CRISPR/Cas9; Genome editing; Livestock; Meganucleases; TALENs; ZFNs.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
References
-
- Bhaya D., Davison M., Barrangou R. CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu. Rev. Genet. 2011;45(1):273–297. - PubMed
-
- Boch J., Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu. Rev. Phytopathol. 2010;48(1):419–436. - PubMed
Publication types
LinkOut - more resources
Full Text Sources

