A Review and Meta-Analysis of Influenza Interactome Studies
- PMID: 35531276
- PMCID: PMC9069142
- DOI: 10.3389/fmicb.2022.869406
A Review and Meta-Analysis of Influenza Interactome Studies
Abstract
Annually, the influenza virus causes 500,000 deaths worldwide. Influenza-associated mortality and morbidity is especially high among the elderly, children, and patients with chronic diseases. While there are antivirals available against influenza, such as neuraminidase inhibitors and adamantanes, there is growing resistance against these drugs. Thus, there is a need for novel antivirals for resistant influenza strains. Host-directed therapies are a potential strategy for influenza as host processes are conserved and are less prone mutations as compared to virus-directed therapies. A literature search was performed for papers that performed viral-host interaction screens and the Reactome pathway database was used for the bioinformatics analysis. A total of 15 studies were curated and 1717 common interactors were uncovered among all these studies. KEGG analysis, Enrichr analysis, STRING interaction analysis was performed on these interactors. Therefore, we have identified novel host pathways that can be targeted for host-directed therapy against influenza in our review.
Keywords: bioinformatics; host-pathogen interactions; influenza; influenza proteins; interactome analysis.
Copyright © 2022 Chua, Cui, Engelberg and Lim.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
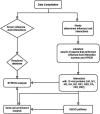

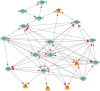
References
Publication types
LinkOut - more resources
Full Text Sources

