The Influence of Host miRNA Binding to RNA Within RNA Viruses on Virus Multiplication
- PMID: 35531344
- PMCID: PMC9069554
- DOI: 10.3389/fcimb.2022.802149
The Influence of Host miRNA Binding to RNA Within RNA Viruses on Virus Multiplication
Abstract
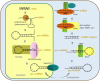
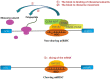
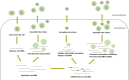
microRNAs (miRNAs), non-coding RNAs about 22 nt long, regulate the post-transcription expression of genes to influence many cellular processes. The expression of host miRNAs is affected by virus invasion, which also affects virus replication. Increasing evidence has demonstrated that miRNA influences RNA virus multiplication by binding directly to the RNA virus genome. Here, the knowledge relating to miRNAs' relationships between host miRNAs and RNA viruses are discussed.
Keywords: RISC complex; RNA virus; argonaute2; flavivirus; miRNA.
Copyright © 2022 Lei, Cheng, Wang and Jia.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Amador-Cañizares Y., Panigrahi M., Huys A., Kunden R. D., Adams H. M., Schinold M. J., et al. (2018). miR-122, Small RNA Annealing and Sequence Mutations Alter the Predicted Structure of the Hepatitis C Virus 5’ UTR RNA to Stabilize and Promote Viral RNA Accumulation. Nucleic Acids Res. 46 (18), 9776–9792. doi: 10.1093/nar/gky662 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

