Profiling senescent cells in human brains reveals neurons with CDKN2D/p19 and tau neuropathology
- PMID: 35531351
- PMCID: PMC9075501
- DOI: 10.1038/s43587-021-00142-3
Profiling senescent cells in human brains reveals neurons with CDKN2D/p19 and tau neuropathology
Abstract
Senescent cells contribute to pathology and dysfunction in animal models1. Their sparse distribution and heterogenous phenotype have presented challenges for detecting them in human tissues. We developed a senescence eigengene approach to identify these rare cells within large, diverse populations of postmortem human brain cells. Eigengenes are useful when no single gene reliably captures a phenotype, like senescence; they also help to reduce noise, which is important in large transcriptomic datasets where subtle signals from low-expressing genes can be lost. Each of our eigengenes detected ~2% senescent cells from a population of ~140,000 single nuclei derived from 76 postmortem human brains with various levels of Alzheimer's disease (AD) pathology. More than 97% of the senescent cells were excitatory neurons and overlapped with tau-containing neurofibrillary tangles (NFTs). Cyclin dependent kinase inhibitor 2D (CDKN2D/p19) was predicted as the most significant contributor to the primary senescence eigengene. RNAscope and immunofluorescence confirmed its elevated expression in AD brain tissue whereby p19-expressing neurons had 1.8-fold larger nuclei and significantly more cells with lipofuscin than p19-negative neurons. These hallmark senescence phenotypes were further elevated in the presence of NFTs. Collectively, CDKN2D/p19-expressing neurons with NFTs represent a unique cellular population in human AD with a senescence phenotype. The eigengenes developed may be useful in future senescence profiling studies as they accurately identified senescent cells in snRNASeq datasets and predicted biomarkers for histological investigation.
Figures

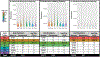
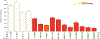

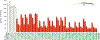





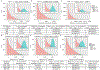
References
Publication types
MeSH terms
Substances
Grants and funding
- P30 AG072975/AG/NIA NIH HHS/United States
- T32 AG021890/AG/NIA NIH HHS/United States
- RF1 AG057440/AG/NIA NIH HHS/United States
- R00 AG061259/AG/NIA NIH HHS/United States
- U01 AG061356/AG/NIA NIH HHS/United States
- R01 AG017917/AG/NIA NIH HHS/United States
- RF1 NS112391/NS/NINDS NIH HHS/United States
- IK2 BX003804/BX/BLRD VA/United States
- R01 AG068293/AG/NIA NIH HHS/United States
- RF1 AG051485/AG/NIA NIH HHS/United States
- R01 AG015819/AG/NIA NIH HHS/United States
- P30 AG062421/AG/NIA NIH HHS/United States
- U01 AG046170/AG/NIA NIH HHS/United States
- R01 AG057907/AG/NIA NIH HHS/United States
- RF1 AG059319/AG/NIA NIH HHS/United States
- R21 AG059176/AG/NIA NIH HHS/United States
- R01 AG057896/AG/NIA NIH HHS/United States
- P30 AG010161/AG/NIA NIH HHS/United States
- K99 AG061259/AG/NIA NIH HHS/United States
- P30 AG066546/AG/NIA NIH HHS/United States
- P30 AG044271/AG/NIA NIH HHS/United States
- RF1 AG057473/AG/NIA NIH HHS/United States
- RF1 AG059082/AG/NIA NIH HHS/United States
- U01 AG052409/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

