Molecular determinants of αVβ5 localization in flat clathrin lattices - role of αVβ5 in cell adhesion and proliferation
- PMID: 35532004
- PMCID: PMC9234671
- DOI: 10.1242/jcs.259465
Molecular determinants of αVβ5 localization in flat clathrin lattices - role of αVβ5 in cell adhesion and proliferation
Abstract
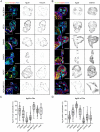
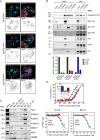
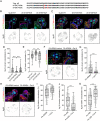
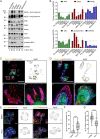
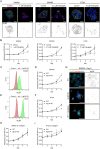
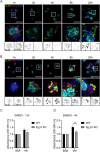
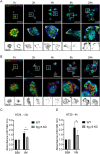
The vitronectin receptor integrin αVβ5 can reside in two distinct adhesion structures - focal adhesions (FAs) and flat clathrin lattices (FCLs). Here, we investigate the mechanism that regulates the subcellular distribution of β5 in keratinocytes and show that β5 has approximately 7- and 5-fold higher affinity for the clathrin adaptors ARH (also known as LDLRAP1) and Numb, respectively, than for the talin 1 (TLN1); all proteins that bind to the membrane-proximal NPxY motif of the β5 cytoplasmic domain. Using mass spectrometry, we identified β5 interactors, including the Rho GEFs p115Rho-GEF and GEF-H1 (also known as ARHGEF1 and ARHGEF2, respectively), and the serine protein kinase MARK2, depletion of which diminishes the clustering of β5 in FCLs. Replacement of two serine residues (S759 and S762) in the β5 cytoplasmic domain with phospho-mimetic glutamate residues causes a shift in the localization of β5 from FAs into FCLs without affecting the interactions with MARK2, p115Rho-GEF or GEF-H1. Instead, we demonstrate that changes in the actomyosin-based cellular contractility by ectopic expression of activated Rho or disruption of microtubules regulates β5 localization. Finally, we present evidence that β5 in either FAs or FCLs functions to promote adhesion to vitronectin, cell spreading, and proliferation.
Keywords: Flat clathrin lattice; Focal adhesion; Integrin; Proliferation; Vitronectin.
© 2022. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interests The authors declare no competing or financial interests.
Figures

Similar articles
-
Mechanisms of integrin αVβ5 clustering in flat clathrin lattices.J Cell Sci. 2018 Nov 5;131(21):jcs221317. doi: 10.1242/jcs.221317. J Cell Sci. 2018. PMID: 30301780
-
The vitronectin receptor associates with clathrin-coated membrane domains via the cytoplasmic domain of its beta5 subunit.J Cell Sci. 1998 Sep;111 ( Pt 18):2729-40. doi: 10.1242/jcs.111.18.2729. J Cell Sci. 1998. PMID: 9718366
-
Reticular adhesions are assembled at flat clathrin lattices and opposed by active integrin α5β1.J Cell Biol. 2023 Aug 7;222(8):e202303107. doi: 10.1083/jcb.202303107. Epub 2023 May 26. J Cell Biol. 2023. PMID: 37233325 Free PMC article.
-
Focal adhesions, reticular adhesions, flat clathrin lattices: what divides them, what unites them?Am J Physiol Cell Physiol. 2025 Jan 1;328(1):C288-C302. doi: 10.1152/ajpcell.00821.2024. Epub 2024 Dec 9. Am J Physiol Cell Physiol. 2025. PMID: 39652817 Review.
-
Integrin-mediated cell adhesion: the cytoskeletal connection.Biochem Soc Symp. 1999;65:79-99. Biochem Soc Symp. 1999. PMID: 10320934 Review.
Cited by
-
The ancestral type of the R-RAS protein has oncogenic potential.Cell Mol Biol Lett. 2024 Feb 21;29(1):27. doi: 10.1186/s11658-024-00546-0. Cell Mol Biol Lett. 2024. PMID: 38383288 Free PMC article.
-
Mechano-regulation by clathrin pit-formation and passive cholesterol-dependent tubules during de-adhesion.Cell Mol Life Sci. 2024 Jan 13;81(1):43. doi: 10.1007/s00018-023-05072-4. Cell Mol Life Sci. 2024. PMID: 38217571 Free PMC article.
-
Canonical and non-canonical integrin-based adhesions dynamically interconvert.Nat Commun. 2024 Mar 7;15(1):2093. doi: 10.1038/s41467-024-46381-x. Nat Commun. 2024. PMID: 38453931 Free PMC article.
-
Flat clathrin lattices are linked to metastatic potential in colorectal cancer.iScience. 2023 Jul 12;26(8):107327. doi: 10.1016/j.isci.2023.107327. eCollection 2023 Aug 18. iScience. 2023. PMID: 37539031 Free PMC article.
-
Dynamics of the formation of flat clathrin lattices in response to growth factor stimulus.bioRxiv [Preprint]. 2025 May 23:2025.05.22.655576. doi: 10.1101/2025.05.22.655576. bioRxiv. 2025. PMID: 40475677 Free PMC article. Preprint.
References
-
- Askham, J. M., Vaughan, K. T., Goodson, H. V. and Morrison, E. E. (2002). Evidence that an interaction between EB1 and p150Glued is required for the formation and maintenance of a radial microtubule array anchored at the centrosome. Mol. Biol. Cell 13, 3627-3645. 10.1091/mbc.e02-01-0061 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

