Serine Protease Inhibitors Restrict Host Susceptibility to SARS-CoV-2 Infections
- PMID: 35532162
- PMCID: PMC9239148
- DOI: 10.1128/mbio.00892-22
Serine Protease Inhibitors Restrict Host Susceptibility to SARS-CoV-2 Infections
Abstract
The coronavirus disease 2019, COVID-19, is a complex disease with a wide range of symptoms from asymptomatic infections to severe acute respiratory syndrome with lethal outcome. Individual factors such as age, sex, and comorbidities increase the risk for severe infections, but other aspects, such as genetic variations, are also likely to affect the susceptibility to SARS-CoV-2 infection and disease severity. Here, we used a human 3D lung cell model based on primary cells derived from multiple donors to identity host factors that regulate SARS-CoV-2 infection. With a transcriptomics-based approach, we found that less susceptible donors show a higher expression level of serine protease inhibitors SERPINA1, SERPINE1, and SERPINE2, identifying variation in cellular serpin levels as restricting host factors for SARS-CoV-2 infection. We pinpoint their antiviral mechanism of action to inhibition of the cellular serine protease, TMPRSS2, thereby preventing cleavage of the viral spike protein and TMPRSS2-mediated entry into the target cells. By means of single-cell RNA sequencing, we further locate the expression of the individual serpins to basal, ciliated, club, and goblet cells. Our results add to the importance of genetic variations as determinants for SARS-CoV-2 susceptibility and suggest that genetic deficiencies of cellular serpins might represent risk factors for severe COVID-19. Our study further highlights TMPRSS2 as a promising target for antiviral intervention and opens the door for the usage of locally administered serpins as a treatment against COVID-19. IMPORTANCE Identification of host factors affecting individual SARS-CoV-2 susceptibility will provide a better understanding of the large variations in disease severity and will identify potential factors that can be used, or targeted, in antiviral drug development. With the use of an advanced lung cell model established from several human donors, we identified cellular protease inhibitors, serpins, as host factors that restrict SARS-CoV-2 infection. The antiviral mechanism was found to be mediated by the inhibition of a serine protease, TMPRSS2, which results in a blockage of viral entry into target cells. Potential treatments with these serpins would not only reduce the overall viral burden in the patients, but also block the infection at an early time point, reducing the risk for the hyperactive immune response common in patients with severe COVID-19.
Keywords: A1AT; ATIII; COVID-19; PAI1; SARS-CoV-2; TMPRSS2; alpha-1-antitrypsin; antithrombin III; plasminogen activator inhibitor 1; serpin.
Conflict of interest statement
The authors declare a conflict of interest. J.T. is employed at Sartorius and A.L. has received unconditional research support from Carl Bennet AB. This does not alter our adherence to mBio policies on sharing data and materials. There are no patents, products in development or marketed products associated with this research to declare.
Figures
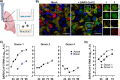
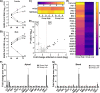
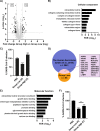
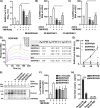
References
-
- Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, Huang H, Zhang L, Zhou X, Du C, Zhang Y, Song J, Wang S, Chao Y, Yang Z, Xu J, Zhou X, Chen D, Xiong W, Xu L, Zhou F, Jiang J, Bai C, Zheng J, Song Y. 2020. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med 180:934–943. doi: 10.1001/jamainternmed.2020.0994. - DOI - PMC - PubMed
-
- Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. 2020. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet 395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. - DOI - PMC - PubMed
-
- Li X, Xu S, Yu M, Wang K, Tao Y, Zhou Y, Shi J, Zhou M, Wu B, Yang Z, Zhang C, Yue J, Zhang Z, Renz H, Liu X, Xie J, Xie M, Zhao J. 2020. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin Immunol 146:110–118. doi: 10.1016/j.jaci.2020.04.006. - DOI - PMC - PubMed
-
- Ellinghaus D, Degenhardt F, Bujanda L, Buti M, Albillos A, Invernizzi P, Fernandez J, Prati D, Baselli G, Asselta R, Grimsrud MM, Milani C, Aziz F, Kassens J, May S, Wendorff M, Wienbrandt L, Uellendahl-Werth F, Zheng T, Yi X, de Pablo R, Chercoles AG, Palom A, Garcia-Fernandez AE, Rodriguez-Frias F, Zanella A, Bandera A, Protti A, Aghemo A, Lleo A, Biondi A, Caballero-Garralda A, Gori A, Tanck A, Carreras Nolla A, Latiano A, Fracanzani AL, Peschuck A, Julia A, Pesenti A, Voza A, Jimenez D, Mateos B, Nafria Jimenez B, Quereda C, Paccapelo C, Gassner C, Angelini C, Cea C, Solier A, et al. 2020. Genomewide association study of severe Covid-19 with respiratory failure. N Engl J Med 383:1522–1534. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

