CRISPR/Cas9 disruption of UGT71L1 in poplar connects salicinoid and salicylic acid metabolism and alters growth and morphology
- PMID: 35532172
- PMCID: PMC9338807
- DOI: 10.1093/plcell/koac135
CRISPR/Cas9 disruption of UGT71L1 in poplar connects salicinoid and salicylic acid metabolism and alters growth and morphology
Abstract
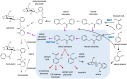
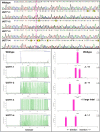
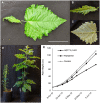
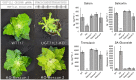
Salicinoids are salicyl alcohol-containing phenolic glycosides with strong antiherbivore effects found only in poplars and willows. Their biosynthesis is poorly understood, but recently a UDP-dependent glycosyltransferase, UGT71L1, was shown to be required for salicinoid biosynthesis in poplar tissue cultures. UGT71L1 specifically glycosylates salicyl benzoate, a proposed salicinoid intermediate. Here, we analyzed transgenic CRISPR/Cas9-generated UGT71L1 knockout plants. Metabolomic analyses revealed substantial reductions in the major salicinoids, confirming the central role of the enzyme in salicinoid biosynthesis. Correspondingly, UGT71L1 knockouts were preferred to wild-type by white-marked tussock moth (Orgyia leucostigma) larvae in bioassays. Greenhouse-grown knockout plants showed substantial growth alterations, with decreased internode length and smaller serrated leaves. Reinserting a functional UGT71L1 gene in a transgenic rescue experiment demonstrated that these effects were due only to the loss of UGT71L1. The knockouts contained elevated salicylate (SA) and jasmonate (JA) concentrations, and also had enhanced expression of SA- and JA-related genes. SA is predicted to be released by UGT71L1 disruption, if salicyl salicylate is a pathway intermediate and UGT71L1 substrate. This idea was supported by showing that salicyl salicylate can be glucosylated by recombinant UGT71L1, providing a potential link of salicinoid metabolism to SA and growth impacts. Connecting this pathway with growth could imply that salicinoids are under additional evolutionary constraints beyond selective pressure by herbivores.
� American Society of Plant Biologists 2022. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures



Comment in
-
CRISPR-Cas9 helps solve a piece of the puzzle of the biosynthesis of salicinoids and suggests a role in the growth-defense trade-off in poplar.Plant Cell. 2022 Jul 30;34(8):2819-2820. doi: 10.1093/plcell/koac157. Plant Cell. 2022. PMID: 35640826 Free PMC article. No abstract available.
Similar articles
-
Discovery of salicyl benzoate UDP-glycosyltransferase, a central enzyme in poplar salicinoid phenolic glycoside biosynthesis.Plant J. 2020 Apr;102(1):99-115. doi: 10.1111/tpj.14615. Epub 2020 Feb 3. Plant J. 2020. PMID: 31736216
-
A willow UDP-glycosyltransferase involved in salicinoid biosynthesis.J Exp Bot. 2021 Feb 27;72(5):1634-1648. doi: 10.1093/jxb/eraa562. J Exp Bot. 2021. PMID: 33249501
-
Functional characterization of two acyltransferases from Populus trichocarpa capable of synthesizing benzyl benzoate and salicyl benzoate, potential intermediates in salicinoid phenolic glycoside biosynthesis.Phytochemistry. 2015 May;113:149-59. doi: 10.1016/j.phytochem.2014.10.018. Epub 2015 Jan 2. Phytochemistry. 2015. PMID: 25561400
-
Jasmonate- and salicylate-mediated plant defense responses to insect herbivores, pathogens and parasitic plants.Pest Manag Sci. 2009 May;65(5):497-503. doi: 10.1002/ps.1714. Pest Manag Sci. 2009. PMID: 19206090 Review.
-
The diversity of salicylic acid biosynthesis and defense signaling in plants: Knowledge gaps and future opportunities.Curr Opin Plant Biol. 2023 Apr;72:102349. doi: 10.1016/j.pbi.2023.102349. Epub 2023 Feb 24. Curr Opin Plant Biol. 2023. PMID: 36842224 Review.
Cited by
-
Poplar leaf bud resin metabolomics: seasonal profiling of leaf bud chemistry in Populus trichocarpa provides insight into resin biosynthesis.Plant Cell Physiol. 2025 Mar 31;66(3):291-303. doi: 10.1093/pcp/pcae149. Plant Cell Physiol. 2025. PMID: 39699046 Free PMC article.
-
Peroxisomal Localization of Benzyl Alcohol O-Benzoyltransferase HSR201 is Mediated by a Non-canonical Peroxisomal Targeting Signal and Required for Salicylic Acid Biosynthesis.Plant Cell Physiol. 2024 Dec 21;65(12):2054-2065. doi: 10.1093/pcp/pcae129. Plant Cell Physiol. 2024. PMID: 39471420 Free PMC article.
-
CRISPR-Cas9 helps solve a piece of the puzzle of the biosynthesis of salicinoids and suggests a role in the growth-defense trade-off in poplar.Plant Cell. 2022 Jul 30;34(8):2819-2820. doi: 10.1093/plcell/koac157. Plant Cell. 2022. PMID: 35640826 Free PMC article. No abstract available.
-
Genomic and transcriptomic analyses reveal polygenic architecture for ecologically important traits in aspen (Populus tremuloides Michx.).Ecol Evol. 2023 Sep 28;13(10):e10541. doi: 10.1002/ece3.10541. eCollection 2023 Oct. Ecol Evol. 2023. PMID: 37780087 Free PMC article.
-
Genome Wide Analysis of Family-1 UDP Glycosyltransferases in Populus trichocarpa Specifies Abiotic Stress Responsive Glycosylation Mechanisms.Genes (Basel). 2022 Sep 13;13(9):1640. doi: 10.3390/genes13091640. Genes (Basel). 2022. PMID: 36140806 Free PMC article.
References
-
- Babst BA, Harding SA, Tsai CJ (2010) Biosynthesis of phenolic glycosides from phenylpropanoid and benzenoid precursors in Populus. J Chem Ecol 36: 286–297 - PubMed
-
- Basey JM, Jenkins SH, Busher PE (1988) Optimal central-place foraging by beavers: tree-size selection in relation to defensive chemicals of quaking aspen. Oecologia 76: 278–282 - PubMed
-
- Brunner AM, Busov VB, Strauss SH (2004) Poplar genome sequence: functional genomics in an ecologically dominant plant species. Trends Plant Sci 9: 49–56 - PubMed
-
- Boeckler G, Gershenzon J, Unsicker S (2011) Phenolic glycosides of the Salicaceae and their role as anti-herbivore defenses. Phytochemistry 72: 1497–1509 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

