Remdesivir and GS-441524 Retain Antiviral Activity against Delta, Omicron, and Other Emergent SARS-CoV-2 Variants
- PMID: 35532238
- PMCID: PMC9211395
- DOI: 10.1128/aac.00222-22
Remdesivir and GS-441524 Retain Antiviral Activity against Delta, Omicron, and Other Emergent SARS-CoV-2 Variants
Abstract
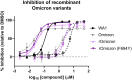
Genetic variation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has resulted in the emergence and rapid spread of multiple variants throughout the pandemic, of which Omicron is currently the predominant variant circulating worldwide. SARS-CoV-2 variants of concern/variants of interest (VOC/VOI) have evidence of increased viral transmission, disease severity, or decreased effectiveness of vaccines and neutralizing antibodies. Remdesivir (RDV [VEKLURY]) is a nucleoside analog prodrug and the first FDA-approved antiviral treatment of COVID-19. Here, we present a comprehensive antiviral activity assessment of RDV and its parent nucleoside, GS-441524, against 10 current and former SARS-CoV-2 VOC/VOI clinical isolates by nucleoprotein enzyme-linked immunosorbent assay (ELISA) and plaque reduction assay. Delta and Omicron variants remained susceptible to RDV and GS-441524, with 50% effective concentration (EC50) values 0.30- to 0.62-fold of those observed against the ancestral WA1 isolate. All other tested variants exhibited EC50 values ranging from 0.13- to 2.3-fold of the observed EC50 values against WA1. Analysis of nearly 6 million publicly available variant isolate sequences confirmed that Nsp12, the RNA-dependent RNA polymerase (RdRp) target of RDV and GS-441524, is highly conserved across variants, with only 2 prevalent changes (P323L and G671S). Using recombinant viruses, both RDV and GS-441524 retained potency against all viruses containing frequent variant substitutions or their combination. Taken together, these results highlight the conserved nature of SARS-CoV-2 Nsp12 and provide evidence of sustained SARS-CoV-2 antiviral activity of RDV and GS-441524 across the tested variants. The observed pan-variant activity of RDV supports its continued use for the treatment of COVID-19 regardless of the SARS-CoV-2 variant.
Keywords: COVID-19; GS-441524; Nsp12; SARS-CoV-2 variants; antiviral agents; remdesivir.
Conflict of interest statement
The authors declare a conflict of interest. The authors affiliated with Gilead Sciences, Inc. are employees of the company and may own company stock. C.K., X.X., and P.-Y.S. received funding from Gilead Sciences, Inc. in support of recombinant virus generation.
Figures



References
-
- Eckerle LD, Becker MM, Halpin RA, Li K, Venter E, Lu X, Scherbakova S, Graham RL, Baric RS, Stockwell TB, Spiro DJ, Denison MR. 2010. Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog 6:e1000896. 10.1371/journal.ppat.1000896. - DOI - PMC - PubMed
-
- Washington NL, Gangavarapu K, Zeller M, Bolze A, Cirulli ET, Schiabor Barrett KM, Larsen BB, Anderson C, White S, Cassens T, Jacobs S, Levan G, Nguyen J, Ramirez JM, III, Rivera-Garcia C, Sandoval E, Wang X, Wong D, Spencer E, Robles-Sikisaka R, Kurzban E, Hughes LD, Deng X, Wang C, Servellita V, Valentine H, De Hoff P, Seaver P, Sathe S, Gietzen K, Sickler B, Antico J, Hoon K, Liu J, Harding A, Bakhtar O, Basler T, Austin B, MacCannell D, Isaksson M, Febbo PG, Becker D, Laurent M, McDonald E, Yeo GW, Knight R, Laurent LC, de Feo E, Worobey M, Chiu CY, et al. 2021. Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell 184:2587–2594.e7. 10.1016/j.cell.2021.03.052. - DOI - PMC - PubMed
-
- Mishra S, Mindermann S, Sharma M, Whittaker C, Mellan TA, Wilton T, Klapsa D, Mate R, Fritzsche M, Zambon M, Ahuja J, Howes A, Miscouridou X, Nason GP, Ratmann O, Semenova E, Leech G, Sandkühler JF, Rogers-Smith C, Vollmer M, Unwin HJT, Gal Y, Chand M, Gandy A, Martin J, Volz E, Ferguson NM, Bhatt S, Brauner JM, Flaxman S, COVID-19 Genomics UK (COG-UK) Consortium . 2021. Changing composition of SARS-CoV-2 lineages and rise of Delta variant in England. EClinicalMedicine 39:101064. 10.1016/j.eclinm.2021.101064. - DOI - PMC - PubMed
-
- Sah P, Vilches TN, Shoukat A, Fitzpatrick MC, Pandey A, Singer BH, Moghadas SM, Galvani AP. 2021. Quantifying the potential dominance of immune-evading SARS-CoV-2 variants in the United States. medRxiv. 10.1101/2021.05.10.21256996. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

