Odorant receptor orthologues in conifer-feeding beetles display conserved responses to ecologically relevant odours
- PMID: 35532927
- PMCID: PMC9321952
- DOI: 10.1111/mec.16494
Odorant receptor orthologues in conifer-feeding beetles display conserved responses to ecologically relevant odours
Abstract
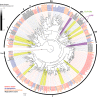
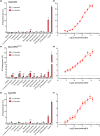
Insects are able to detect a plethora of olfactory cues using a divergent family of odorant receptors (ORs). Despite the divergent nature of this family, related species frequently express several evolutionarily conserved OR orthologues. In the largest order of insects, Coleoptera, it remains unknown whether OR orthologues have conserved or divergent functions in different species. Using HEK293 cells, we addressed this question through functional characterization of two groups of OR orthologues in three species of the Curculionidae (weevil) family, the conifer-feeding bark beetles Ips typographus L. ("Ityp") and Dendroctonus ponderosae Hopkins ("Dpon") (Scolytinae), and the pine weevil Hylobius abietis L. ("Habi"; Molytinae). The ORs of H. abietis were annotated from antennal transcriptomes. The results show highly conserved response specificities, with one group of orthologues (HabiOR3/DponOR8/ItypOR6) responding exclusively to 2-phenylethanol (2-PE), and the other group (HabiOR4/DponOR9/ItypOR5) responding to angiosperm green leaf volatiles (GLVs). Both groups of orthologues belong to the coleopteran OR subfamily 2B, and share a common ancestor with OR5 in the cerambycid Megacyllene caryae, also tuned to 2-PE, suggesting a shared evolutionary history of 2-PE receptors across two beetle superfamilies. The detected compounds are ecologically relevant for conifer-feeding curculionids, and are probably linked to fitness, with GLVs being used to avoid angiosperm nonhost plants, and 2-PE being important for intraspecific communication and/or playing a putative role in beetle-microbe symbioses. To our knowledge, this study is the first to reveal evolutionary conservation of OR functions across several beetle species and hence sheds new light on the functional evolution of insect ORs.
Keywords: Coleoptera; Curculionidae; HEK293 cells; de-orphanization; evolutionary conservation; functional characterization.
© 2022 The Authors. Molecular Ecology published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflicts of interest to disclose.
Figures

References
-
- Andersson, M. N. , Grosse‐Wilde, E. , Keeling, C. I. , Bengtsson, J. M. , Yuen, M. M. , Li, M. , Hillbur, Y. , Bohlmann, J. , Hansson, B. S. , & Schlyter, F. (2013). Antennal transcriptome analysis of the chemosensory gene families in the tree killing bark beetles, Ips typographus and Dendroctonus ponderosae (Coleoptera: Curculionidae: Scolytinae). BMC Genomics, 14(1), 198. 10.1186/1471-2164-14-198 - DOI - PMC - PubMed
-
- Andersson, M. N. , Larsson, M. C. , Svensson, G. P. , Birgersson, G. , Rundlöf, M. , Lundin, O. , Lankinen, Å. , & Anderbrant, O. (2012). Characterization of olfactory sensory neurons in the white clover seed weevil, Apion fulvipes (Coleoptera: Apionidae). Journal of Insect Physiology, 58, 1325–1333. 10.1016/j.jinsphys.2012.07.006 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

