On the Utility of ToxCast-Based Predictive Models to Evaluate Potential Metabolic Disruption by Environmental Chemicals
- PMID: 35533074
- PMCID: PMC9084331
- DOI: 10.1289/EHP6779
On the Utility of ToxCast-Based Predictive Models to Evaluate Potential Metabolic Disruption by Environmental Chemicals
Abstract
Background: Research suggests environmental contaminants can impact metabolic health; however, high costs prohibit in vivo screening of putative metabolic disruptors. High-throughput screening programs, such as ToxCast, hold promise to reduce testing gaps and prioritize higher-order (in vivo) testing.
Objectives: We sought to a) examine the concordance of in vitro testing in 3T3-L1 cells to a targeted literature review for 38 semivolatile environmental chemicals, and b) assess the predictive utility of various expert models using ToxCast data against the set of 38 reference chemicals.
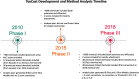
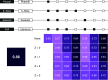
Methods: Using a set of 38 chemicals with previously published results in 3T3-L1 cells, we performed a metabolism-targeted literature review to determine consensus activity determinations. To assess ToxCast predictive utility, we used two published ToxPi models: a) the 8-Slice model published by Janesick et al. (2016) and b) the 5-Slice model published by Auerbach et al. (2016). We examined the performance of the two models against the Janesick in vitro results and our own 38-chemical reference set. We further evaluated the predictive performance of various modifications to these models using cytotoxicity filtering approaches and validated our best-performing model with new chemical testing in 3T3-L1 cells.
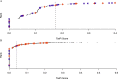
Results: The literature review revealed relevant publications for 30 out of the 38 chemicals (the remaining 8 chemicals were only examined in our previous 3T3-L1 testing). We observed a balanced accuracy (average of sensitivity and specificity) of 0.86 comparing our previous in vitro results to the literature-derived calls. ToxPi models provided balanced accuracies ranging from 0.55 to 0.88, depending on the model specifications and reference set. Validation chemical testing correctly predicted 29 of 30 chemicals as per 3T3-L1 testing, suggesting good adipogenic prediction performance for our best adapted model.
Discussion: Using the most recent ToxCast data and an updated ToxPi model, we found ToxCast performed similarly to that of our own 3T3-L1 testing in predicting consensus calls. Furthermore, we provide the full ranked list of largely untested chemicals with ToxPi scores that predict adipogenic activity and that require further investigation. https://doi.org/10.1289/EHP6779.
Figures

References
-
- Angle BM, Do RP, Ponzi D, Stahlhut RW, Drury BE, Nagel SC, et al. 2013. Metabolic disruption in male mice due to fetal exposure to low but not high doses of bisphenol A (BPA): evidence for effects on body weight, food intake, adipocytes, leptin, adiponectin, insulin and glucose regulation. Reprod Toxicol 42:13, PMID: , 10.1016/j.reprotox.2013.07.017. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
