Central Visual Function and Genotype-Phenotype Correlations in PDE6A-Associated Retinitis Pigmentosa
- PMID: 35533076
- PMCID: PMC9106976
- DOI: 10.1167/iovs.63.5.9
Central Visual Function and Genotype-Phenotype Correlations in PDE6A-Associated Retinitis Pigmentosa
Abstract
Purpose: Autosomal recessive retinitis pigmentosa (arRP) can be caused by mutations in the phosphodiesterase 6A (PDE6A) gene. Here, we describe the natural course of disease progression with respect to central retinal function (i.e., visual acuity, contrast sensitivity, and color vision) and establish a detailed genotype--phenotype correlation.
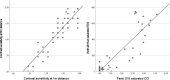
Methods: Forty-four patients (26 females; mean age ± SD, 43 ± 13 years) with a confirmed genetic diagnosis of PDE6A-associated arRP underwent comprehensive ophthalmological examinations including best-corrected visual acuity (BCVA) with Early Treatment Diabetic Retinopathy Study charts, contrast sensitivity (CS) with Pelli-Robson charts at distances of 3 m and 1 m, and color vision testing using Roth 28-Hue and Panel D-15 saturated color cups.
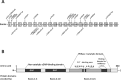
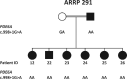
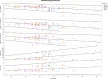
Results: The most frequently observed variants were c.998+1G>A/p.?, c.304C>A/p.R102S, and c.2053G>A/p.V685M. Central retinal function in patients homozygous for variant c.304C>A/p.R102S was better when compared to patients homozygous for variant c.998+1G>A/p.?, although the former were older at baseline. Central retinal function was similar in patients homozygous for variant c.304C>A/p.R102S and patients heterozygous for variants c.304C>A/p.R102S and c.2053G>A/p.V685M, although the latter were younger at baseline. Annual decline rates in central retinal function were small.
Conclusions: We conclude that the severity of the different disease-causing PDE6A mutations in humans with respect to central visual function may be ranked as follows: c.2053G>A/p.V685M in homozygous state (most severe) > c.998+1G>A/p.? in homozygous state > c.304C>A/p.R102S and c.2053G>A/p.V685M in compound-heterozygous state > c.304C>A/p.R102S in homozygous state (mildest). The assessment of treatment efficacy in interventional trials will remain challenging due to small annual decline rates in central retinal function.
Conflict of interest statement
Disclosure:
Figures

References
-
- Pagon RA. Retinitis pigmentosa. Surv Ophthalmol. 1988; 33(3): 137–177. - PubMed
-
- Retinal Information Network. RetNet: Retinal Information Network. Available at: https://sph.uth.edu/RETNET/. Accessed April 28, 2022.
-
- Tsang SH, Sharma T. Retinitis pigmentosa (non-syndromic). Adv Exp Med Biol. 2018; 1085: 125–130. - PubMed
-
- Huang SH, Pittler SJ, Huang X, Oliveira L, Berson EL, Dryja TP. Autosomal recessive retinitis pigmentosa caused by mutations in the alpha subunit of rod cGMP phosphodiesterase. Nat Genet. 1995; 11(4): 468–471. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

