TRIM25 and ZAP target the Ebola virus ribonucleoprotein complex to mediate interferon-induced restriction
- PMID: 35533151
- PMCID: PMC9119685
- DOI: 10.1371/journal.ppat.1010530
TRIM25 and ZAP target the Ebola virus ribonucleoprotein complex to mediate interferon-induced restriction
Abstract
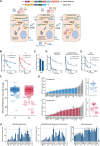
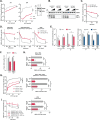
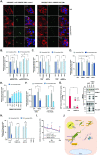
Ebola virus (EBOV) causes highly pathogenic disease in primates. Through screening a library of human interferon-stimulated genes (ISGs), we identified TRIM25 as a potent inhibitor of EBOV transcription-and-replication-competent virus-like particle (trVLP) propagation. TRIM25 overexpression inhibited the accumulation of viral genomic and messenger RNAs independently of the RNA sensor RIG-I or secondary proinflammatory gene expression. Deletion of TRIM25 strongly attenuated the sensitivity of trVLPs to inhibition by type-I interferon. The antiviral activity of TRIM25 required ZAP and the effect of type-I interferon was modulated by the CpG dinucleotide content of the viral genome. We find that TRIM25 interacts with the EBOV vRNP, resulting in its autoubiquitination and ubiquitination of the viral nucleoprotein (NP). TRIM25 is recruited to incoming vRNPs shortly after cell entry and leads to dissociation of NP from the vRNA. We propose that TRIM25 targets the EBOV vRNP, exposing CpG-rich viral RNA species to restriction by ZAP.
Conflict of interest statement
The authors have declared no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

