Influence of confinement on the spreading of bacterial populations
- PMID: 35533196
- PMCID: PMC9119553
- DOI: 10.1371/journal.pcbi.1010063
Influence of confinement on the spreading of bacterial populations
Abstract
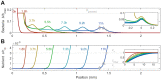
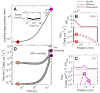
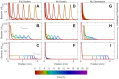
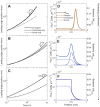
The spreading of bacterial populations is central to processes in agriculture, the environment, and medicine. However, existing models of spreading typically focus on cells in unconfined settings-despite the fact that many bacteria inhabit complex and crowded environments, such as soils, sediments, and biological tissues/gels, in which solid obstacles confine the cells and thereby strongly regulate population spreading. Here, we develop an extended version of the classic Keller-Segel model of bacterial spreading via motility that also incorporates cellular growth and division, and explicitly considers the influence of confinement in promoting both cell-solid and cell-cell collisions. Numerical simulations of this extended model demonstrate how confinement fundamentally alters the dynamics and morphology of spreading bacterial populations, in good agreement with recent experimental results. In particular, with increasing confinement, we find that cell-cell collisions increasingly hinder the initial formation and the long-time propagation speed of chemotactic pulses. Moreover, also with increasing confinement, we find that cellular growth and division plays an increasingly dominant role in driving population spreading-eventually leading to a transition from chemotactic spreading to growth-driven spreading via a slower, jammed front. This work thus provides a theoretical foundation for further investigations of the influence of confinement on bacterial spreading. More broadly, these results help to provide a framework to predict and control the dynamics of bacterial populations in complex and crowded environments.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Chaban B, Hughes HV, Beeby M. The flagellum in bacterial pathogens: for motility and a whole lot more. In: Seminars in cell and developmental biology. vol. 46. Elsevier; 2015. p. 91–103. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

