A randomization-based causal inference framework for uncovering environmental exposure effects on human gut microbiota
- PMID: 35533202
- PMCID: PMC9129050
- DOI: 10.1371/journal.pcbi.1010044
A randomization-based causal inference framework for uncovering environmental exposure effects on human gut microbiota
Abstract
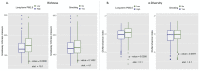
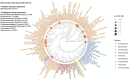
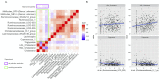
Statistical analysis of microbial genomic data within epidemiological cohort studies holds the promise to assess the influence of environmental exposures on both the host and the host-associated microbiome. However, the observational character of prospective cohort data and the intricate characteristics of microbiome data make it challenging to discover causal associations between environment and microbiome. Here, we introduce a causal inference framework based on the Rubin Causal Model that can help scientists to investigate such environment-host microbiome relationships, to capitalize on existing, possibly powerful, test statistics, and test plausible sharp null hypotheses. Using data from the German KORA cohort study, we illustrate our framework by designing two hypothetical randomized experiments with interventions of (i) air pollution reduction and (ii) smoking prevention. We study the effects of these interventions on the human gut microbiome by testing shifts in microbial diversity, changes in individual microbial abundances, and microbial network wiring between groups of matched subjects via randomization-based inference. In the smoking prevention scenario, we identify a small interconnected group of taxa worth further scrutiny, including Christensenellaceae and Ruminococcaceae genera, that have been previously associated with blood metabolite changes. These findings demonstrate that our framework may uncover potentially causal links between environmental exposure and the gut microbiome from observational data. We anticipate the present statistical framework to be a good starting point for further discoveries on the role of the gut microbiome in environmental health.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(10):3698–3703. doi: 10.1073/pnas.0812874106 - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

