Stromal p53 Regulates Breast Cancer Development, the Immune Landscape, and Survival in an Oncogene-Specific Manner
- PMID: 35533313
- PMCID: PMC9357052
- DOI: 10.1158/1541-7786.MCR-21-0960
Stromal p53 Regulates Breast Cancer Development, the Immune Landscape, and Survival in an Oncogene-Specific Manner
Abstract
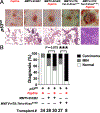
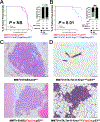
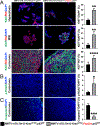
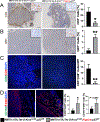
Coevolution of tumor cells and adjacent stromal elements is a key feature during tumor progression; however, the precise regulatory mechanisms during this process remain unknown. Here, we show stromal p53 loss enhances oncogenic KrasG12D, but not ErbB2, driven tumorigenesis in murine mammary epithelia. Stroma-specific p53 deletion increases both epithelial and fibroblast proliferation in mammary glands bearing the KrasG12D oncogene in epithelia, while concurrently increasing DNA damage and/or DNA replication stress and decreasing apoptosis in the tumor cells proper. Normal epithelia was not affected by stromal p53 deletion. Tumors with p53-null stroma had a significant decrease in total, cytotoxic, and regulatory T cells; however, there was a significant increase in myeloid-derived suppressor cells, total macrophages, and M2-polarized tumor-associated macrophages, with no impact on angiogenesis or connective tissue deposition. Stroma-specific p53 deletion reprogrammed gene expression in both fibroblasts and adjacent epithelium, with p53 targets and chemokine receptors/chemokine signaling pathways in fibroblasts and DNA replication, DNA damage repair, and apoptosis in epithelia being the most significantly impacted biological processes. A gene cluster in p53-deficient mouse fibroblasts was negatively associated with patient survival when compared with two independent datasets. In summary, stroma-specific p53 loss promotes mammary tumorigenesis in an oncogene-specific manner, influences the tumor immune landscape, and ultimately impacts patient survival.
Implications: Expression of the p53 tumor suppressor in breast cancer tumor stroma regulates tumorigenesis in an oncogene-specific manner, influences the tumor immune landscape, and ultimately impacts patient survival.
©2022 American Association for Cancer Research.
Conflict of interest statement
Figures


Similar articles
-
Interleukin-1β promotes ovarian tumorigenesis through a p53/NF-κB-mediated inflammatory response in stromal fibroblasts.Neoplasia. 2013 Apr;15(4):409-20. doi: 10.1593/neo.121228. Neoplasia. 2013. PMID: 23555186 Free PMC article.
-
Cyclophilin D counteracts P53-mediated growth arrest and promotes Ras tumorigenesis.Oncogene. 2016 Sep 29;35(39):5132-43. doi: 10.1038/onc.2016.42. Epub 2016 Mar 14. Oncogene. 2016. PMID: 26973251
-
Mutations in bone marrow-derived stromal stem cells unmask latent malignancy.Stem Cells Dev. 2010 Aug;19(8):1153-66. doi: 10.1089/scd.2009.0439. Stem Cells Dev. 2010. PMID: 20199238 Free PMC article.
-
Role of proto-oncogene activation in carcinogenesis.Environ Health Perspect. 1992 Nov;98:13-24. doi: 10.1289/ehp.929813. Environ Health Perspect. 1992. PMID: 1486840 Free PMC article. Review.
-
Activation of p53 by oncogenes.Endocr Relat Cancer. 1999 Mar;6(1):45-8. doi: 10.1677/erc.0.0060045. Endocr Relat Cancer. 1999. PMID: 10732786 Review.
Cited by
-
Impact of NDUFAF6 on breast cancer prognosis: linking mitochondrial regulation to immune response and PD-L1 expression.Cancer Cell Int. 2024 Mar 8;24(1):99. doi: 10.1186/s12935-024-03244-1. Cancer Cell Int. 2024. PMID: 38459583 Free PMC article.
-
The Tumour Glyco-Code: Sialylation as a Mediator of Stromal Cell Immunosuppression in the Tumour Microenvironment.Eur J Immunol. 2025 Jul;55(7):e70000. doi: 10.1002/eji.70000. Eur J Immunol. 2025. PMID: 40667828 Free PMC article. Review.
-
The effects of dendritic cell-based vaccines in the tumor microenvironment: Impact on myeloid-derived suppressor cells.Front Immunol. 2022 Nov 15;13:1050484. doi: 10.3389/fimmu.2022.1050484. eCollection 2022. Front Immunol. 2022. PMID: 36458011 Free PMC article. Review.
-
Tumor suppressor genes in the tumor microenvironment.Dis Model Mech. 2025 Mar 1;18(3):dmm052049. doi: 10.1242/dmm.052049. Epub 2025 Mar 20. Dis Model Mech. 2025. PMID: 40110599 Free PMC article. Review.
References
-
- Kalluri R The biology and function of fibroblasts in cancer. Nat Rev Cancer 2016;16:582–98 - PubMed
-
- Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ 2006;13:1027–36 - PubMed
-
- Hartmaier RJ, Albacker LA, Chmielecki J, Bailey M, He J, Goldberg ME, et al. High-Throughput Genomic Profiling of Adult Solid Tumors Reveals Novel Insights into Cancer Pathogenesis. Cancer Res 2017;77:2464–75 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

