Network for network concept offers new insights into host- SARS-CoV-2 protein interactions and potential novel targets for developing antiviral drugs
- PMID: 35533462
- PMCID: PMC9055686
- DOI: 10.1016/j.compbiomed.2022.105575
Network for network concept offers new insights into host- SARS-CoV-2 protein interactions and potential novel targets for developing antiviral drugs
Abstract
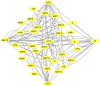
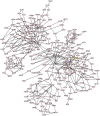
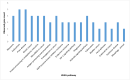
SARS-CoV-2, the causal agent of COVID-19, is primarily a pulmonary virus that can directly or indirectly infect several organs. Despite many studies carried out during the current COVID-19 pandemic, some pathological features of SARS-CoV-2 have remained unclear. It has been recently attempted to address the current knowledge gaps on the viral pathogenicity and pathological mechanisms via cellular-level tropism of SARS-CoV-2 using human proteomics, visualization of virus-host protein-protein interactions (PPIs), and enrichment analysis of experimental results. The synergistic use of models and methods that rely on graph theory has enabled the visualization and analysis of the molecular context of virus/host PPIs. We review current knowledge on the SARS-COV-2/host interactome cascade involved in the viral pathogenicity through the graph theory concept and highlight the hub proteins in the intra-viral network that create a subnet with a small number of host central proteins, leading to cell disintegration and infectivity. Then we discuss the putative principle of the "gene-for-gene and "network for network" concepts as platforms for future directions toward designing efficient anti-viral therapies.
Keywords: Gene network; Protein-protein interactions; SARS-CoV-2; Virus-host interactome.
Copyright © 2022 Elsevier Ltd. All rights reserved.
Figures


References
-
- Figures K. 2022. Weekly Operational Update on COVID-19; pp. 1–11.
-
- WHO . vols. 1–23. World Heal. Organ; 2021. https://www.who.int/publications/m/item/covid-19-weekly-epidemiological-... (COVID-19 Weekly Epidemiological Update).
-
- WHO . vol. 19. 2021. https://covid19.who.int/ (Weekly Epidemiological Update).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

