Analysis of a SARS-CoV-2 convalescent cohort identified a common strategy for escape of vaccine-induced anti-RBD antibodies by Beta and Omicron variants
- PMID: 35533497
- PMCID: PMC9073271
- DOI: 10.1016/j.ebiom.2022.104025
Analysis of a SARS-CoV-2 convalescent cohort identified a common strategy for escape of vaccine-induced anti-RBD antibodies by Beta and Omicron variants
Abstract
Background: Evolutionary pressure has led to the emergence of SARS-CoV-2 variants, with the most recent Omicron variant containing an unparalleled 30 mutations in the spike protein. Many of these mutations are expected to increase immune evasion, thus making breakthrough cases and re-infection more common.
Methods: From June 2020 to December 2021 serial blood samples (initial post recovery, 6 months, 12 months) were collected from a COVID-19 convalescent cohort in Boston, MA. Plasma was isolated for use in Mesoscale Discovery based antibody binding assays. Unvaccinated donors or those vaccinated prior to the primary blood draw were excluded from this analysis, as were those who did not have at least two blood draws. Wilcoxon signed rank tests were used to compare pre- and post-vaccination titers and antibody response against different variants, while McNemar tests were used to compare the proportions of achieving ≥ 4 fold increases against different variants.
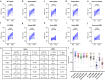
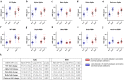
Findings: Forty-eight COVID convalescent donors with post-infection vaccination (hybrid immunity) were studied to evaluate the levels of cross-reactive antibodies pre- and post- vaccination against various SARS-CoV-2 Spike and receptor binding domain (RBD) proteins. Vaccination with BNT162b2, mRNA-1273 or Ad26.COV2.S led to a 6·3 to 7·8 fold increase in anti-Spike antibody titers and a 7·0 to 7·4 fold increase in anti-WT, Alpha and Delta RBD antibody. However, a lower response was observed for Beta and Omicron RBDs with only 7/48 (15%) and 15/48 (31%) donors having a ≥4 fold increase in post-vaccination titers against Beta and Omicron RBDs. Structural analysis of the Beta and Omicron RBDs reveal a shared immune escape strategy involving residues K417-E484-N501 that is exploited by these variants of concern.
Interpretation: Through mutations of the K417-E484-N501 triad, SARS-CoV-2 has evolved to evade neutralization by the class I/II anti-RBD antibody fraction of hybrid immunity plasma as the polyclonal antibody response post-vaccination shows limitations in the ability to solve the structural requirements to bind the mutant RBDs.
Funding: Massachusetts Consortium on Pathogen Readiness (280870.5116709.0016) and the National Institute of Allergy and Infectious Diseases (1R01AI161152-01A1).
Keywords: COVID-19; Hybrid immunity; Neutralization escape mutations; Omicron variant; SARS-CoV-2; Vaccine induced antibody titers.
Copyright © 2022 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Pre-Omicron Vaccine Breakthrough Infection Induces Superior Cross-Neutralization against SARS-CoV-2 Omicron BA.1 Compared to Infection Alone.Int J Mol Sci. 2022 Jul 12;23(14):7675. doi: 10.3390/ijms23147675. Int J Mol Sci. 2022. PMID: 35887023 Free PMC article.
-
A mosaic-type trimeric RBD-based COVID-19 vaccine candidate induces potent neutralization against Omicron and other SARS-CoV-2 variants.Elife. 2022 Aug 25;11:e78633. doi: 10.7554/eLife.78633. Elife. 2022. PMID: 36004719 Free PMC article.
-
SARS-CoV-2 Infection-and mRNA Vaccine-induced Humoral Immunity among Schoolchildren in Hawassa, Ethiopia.Front Immunol. 2023 Jun 15;14:1163688. doi: 10.3389/fimmu.2023.1163688. eCollection 2023. Front Immunol. 2023. PMID: 37398668 Free PMC article.
-
The Biological Functions and Clinical Significance of SARS-CoV-2 Variants of Corcern.Front Med (Lausanne). 2022 May 20;9:849217. doi: 10.3389/fmed.2022.849217. eCollection 2022. Front Med (Lausanne). 2022. PMID: 35669924 Free PMC article. Review.
-
Analysis of anti-SARS-CoV-2 Omicron-neutralizing antibody titers in different vaccinated and unvaccinated convalescent plasma sources.Nat Commun. 2022 Oct 29;13(1):6478. doi: 10.1038/s41467-022-33864-y. Nat Commun. 2022. PMID: 36309490 Free PMC article.
Cited by
-
SARS-CoV-2 Antibody Effectiveness Is Influenced by Non-Epitope Mutation/Binding-Induced Denaturation of the Epitope 3D Architecture.Pathogens. 2022 Nov 29;11(12):1437. doi: 10.3390/pathogens11121437. Pathogens. 2022. PMID: 36558771 Free PMC article.
-
Intersecting SARS-CoV-2 spike mutations and global vaccine efficacy against COVID-19.Front Immunol. 2025 Mar 7;16:1435873. doi: 10.3389/fimmu.2025.1435873. eCollection 2025. Front Immunol. 2025. PMID: 40124365 Free PMC article.
-
Immune Evasion of SARS-CoV-2 Omicron Subvariants.Vaccines (Basel). 2022 Sep 16;10(9):1545. doi: 10.3390/vaccines10091545. Vaccines (Basel). 2022. PMID: 36146623 Free PMC article.
-
Longitudinal Serological Surveillance for COVID-19 Antibodies after Infection and Vaccination.Microbiol Spectr. 2022 Oct 26;10(5):e0202622. doi: 10.1128/spectrum.02026-22. Epub 2022 Sep 19. Microbiol Spectr. 2022. PMID: 36121258 Free PMC article.
-
The evolution of SARS-CoV-2.Nat Rev Microbiol. 2023 Jun;21(6):361-379. doi: 10.1038/s41579-023-00878-2. Epub 2023 Apr 5. Nat Rev Microbiol. 2023. PMID: 37020110 Review.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

