Novel POLE mutations identified in patients with IMAGE-I syndrome cause aberrant subcellular localisation and protein degradation in the nucleus
- PMID: 35534205
- PMCID: PMC9613869
- DOI: 10.1136/jmedgenet-2021-108300
Novel POLE mutations identified in patients with IMAGE-I syndrome cause aberrant subcellular localisation and protein degradation in the nucleus
Abstract
Background: DNA replisome is a molecular complex that plays indispensable roles in normal DNA replication. IMAGE-I syndrome is a DNA replisome-associated genetic disease caused by biallelic mutations in the gene encoding DNA polymerase epsilon catalytic subunit 1 (POLE). However, the underlying molecular mechanisms remain largely unresolved.
Methods: The clinical manifestations in two patients with IMAGE-I syndrome were characterised. Whole-exome sequencing was performed and altered mRNA splicing and protein levels of POLE were determined. Subcellular localisation, cell cycle analysis and DNA replication stress were assessed using fibroblasts and peripheral blood from the patients and transfected cell lines to determine the functional significance of POLE mutations.
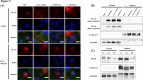
Results: Both patients presented with growth retardation, adrenal insufficiency, immunodeficiency and complicated diffuse large B-cell lymphoma. We identified three novel POLE mutations: namely, a deep intronic mutation, c.1226+234G>A, common in both patients, and missense (c.2593T>G) and in-frame deletion (c.711_713del) mutations in each patient. The unique deep intronic mutation produced aberrantly spliced mRNAs. All mutants showed significantly reduced, but not null, protein levels. Notably, the mutants showed severely diminished nuclear localisation, which was rescued by proteasome inhibitor treatment. Functional analysis revealed impairment of cell cycle progression and increase in the expression of phospho-H2A histone family member X in both patients.
Conclusion: These findings provide new insights regarding the mechanism via which POLE mutants are highly susceptible to proteasome-dependent degradation in the nucleus, resulting in impaired DNA replication and cell cycle progression, a characteristic of DNA replisome-associated diseases.
Keywords: DNA replication; immune system diseases; mutation.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures




References
-
- Cottineau J, Kottemann MC, Lach FP, Kang Y-H, Vély F, Deenick EK, Lazarov T, Gineau L, Wang Y, Farina A, Chansel M, Lorenzo L, Piperoglou C, Ma CS, Nitschke P, Belkadi A, Itan Y, Boisson B, Jabot-Hanin F, Picard C, Bustamante J, Eidenschenk C, Boucherit S, Aladjidi N, Lacombe D, Barat P, Qasim W, Hurst JA, Pollard AJ, Uhlig HH, Fieschi C, Michon J, Bermudez VP, Abel L, de Villartay J-P, Geissmann F, Tangye SG, Hurwitz J, Vivier E, Casanova J-L, Smogorzewska A, Jouanguy E. Inherited GINS1 deficiency underlies growth retardation along with neutropenia and NK cell deficiency. J Clin Invest 2017;127:1991–2006. 10.1172/JCI90727 - DOI - PMC - PubMed
-
- Narumi S, Amano N, Ishii T, Katsumata N, Muroya K, Adachi M, Toyoshima K, Tanaka Y, Fukuzawa R, Miyako K, Kinjo S, Ohga S, Ihara K, Inoue H, Kinjo T, Hara T, Kohno M, Yamada S, Urano H, Kitagawa Y, Tsugawa K, Higa A, Miyawaki M, Okutani T, Kizaki Z, Hamada H, Kihara M, Shiga K, Yamaguchi T, Kenmochi M, Kitajima H, Fukami M, Shimizu A, Kudoh J, Shibata S, Okano H, Miyake N, Matsumoto N, Hasegawa T. Samd9 mutations cause a novel multisystem disorder, mirage syndrome, and are associated with loss of chromosome 7. Nat Genet 2016;48:792–7. 10.1038/ng.3569 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
